Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
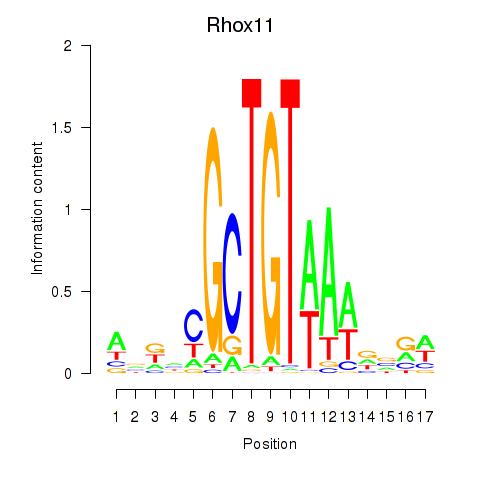
Results for Rhox11
Z-value: 0.83
Transcription factors associated with Rhox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox11
|
ENSRNOG00000028044 | reproductive homeobox 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rhox11 | rn6_v1_chrX_+_124228270_124228270 | -0.02 | 7.1e-01 | Click! |
Activity profile of Rhox11 motif
Sorted Z-values of Rhox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_72272248 | 37.40 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr18_+_70548607 | 32.45 |
ENSRNOT00000085318
|
Myo5b
|
myosin Vb |
| chr4_+_168832910 | 25.14 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr5_+_119727839 | 18.17 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr14_+_109533792 | 16.72 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr5_-_138462864 | 16.58 |
ENSRNOT00000011325
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr20_+_46044892 | 15.51 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr9_-_30321323 | 15.28 |
ENSRNOT00000018234
|
Fam135a
|
family with sequence similarity 135, member A |
| chr13_+_47454591 | 14.83 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr6_-_49324450 | 14.65 |
ENSRNOT00000066904
|
Sntg2
|
syntrophin, gamma 2 |
| chr8_-_14880644 | 14.49 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr1_+_33910912 | 13.97 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr6_-_88917070 | 13.53 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chrX_+_35599258 | 13.08 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr2_-_183213228 | 12.78 |
ENSRNOT00000067618
|
Trim2
|
tripartite motif-containing 2 |
| chr13_-_80862963 | 12.65 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr14_-_77810147 | 11.80 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chrX_-_40086870 | 11.55 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr9_-_91683468 | 11.15 |
ENSRNOT00000041550
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr3_+_56355431 | 11.14 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr5_+_74727494 | 10.71 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr19_+_38422164 | 10.65 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr2_+_252090669 | 10.29 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr2_+_55747318 | 10.25 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chr2_+_3662763 | 10.03 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr4_+_174692331 | 9.72 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr2_-_18531210 | 9.68 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr1_-_52962388 | 9.56 |
ENSRNOT00000033685
|
T2
|
brachyury 2 |
| chr3_+_2262253 | 9.45 |
ENSRNOT00000042100
ENSRNOT00000061303 ENSRNOT00000048137 ENSRNOT00000048353 ENSRNOT00000012129 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr4_-_14490446 | 9.43 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr1_+_156552328 | 8.50 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr8_-_53816447 | 8.41 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr18_+_30527705 | 8.39 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr3_+_35175313 | 8.38 |
ENSRNOT00000081744
|
Kif5c
|
kinesin family member 5C |
| chr17_+_89452814 | 8.26 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr20_+_26893016 | 7.96 |
ENSRNOT00000082430
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr1_+_166428761 | 7.95 |
ENSRNOT00000085555
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr11_-_35749464 | 7.78 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chrX_+_908044 | 7.77 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr1_+_234252757 | 7.77 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr3_-_52510507 | 7.17 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr1_-_47477524 | 6.80 |
ENSRNOT00000067916
|
Rsph3
|
radial spoke 3 homolog |
| chr7_+_71572941 | 6.77 |
ENSRNOT00000007255
|
Sdc2
|
syndecan 2 |
| chr1_-_81846804 | 6.20 |
ENSRNOT00000027435
|
Rabac1
|
Rab acceptor 1 |
| chr1_-_252808380 | 6.18 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr16_-_7406951 | 5.75 |
ENSRNOT00000066508
|
Dnah1
|
dynein, axonemal, heavy chain 1 |
| chr8_-_84506328 | 5.74 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr6_-_135939534 | 5.66 |
ENSRNOT00000052237
|
Diras3
|
DIRAS family GTPase 3 |
| chr7_+_15422479 | 5.44 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr2_-_84436912 | 5.23 |
ENSRNOT00000059565
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr16_+_69048730 | 5.10 |
ENSRNOT00000086082
ENSRNOT00000078128 |
Rab11fip1
|
RAB11 family interacting protein 1 |
| chr3_+_8547349 | 5.08 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr1_-_146289465 | 4.97 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr5_-_153840178 | 4.95 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr1_-_266914093 | 4.69 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr6_-_3395954 | 4.68 |
ENSRNOT00000078044
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr9_-_78969013 | 4.66 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr9_+_30419001 | 4.63 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chr2_-_84437084 | 4.42 |
ENSRNOT00000077598
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr14_+_113867209 | 4.42 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr7_-_135850542 | 4.42 |
ENSRNOT00000038032
|
Twf1
|
twinfilin actin-binding protein 1 |
| chr4_-_165192647 | 4.08 |
ENSRNOT00000086461
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr6_+_8669722 | 3.99 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr3_+_153491151 | 3.96 |
ENSRNOT00000047047
|
Manbal
|
mannosidase beta like |
| chr5_+_104941066 | 3.86 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr13_+_87986240 | 3.45 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_-_120373500 | 3.37 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chr11_+_71151132 | 3.33 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr3_-_21027947 | 3.32 |
ENSRNOT00000051973
|
Olr421
|
olfactory receptor 421 |
| chr5_-_107505454 | 3.17 |
ENSRNOT00000051752
|
Ifna11
|
interferon alpha family, gene 11 |
| chr9_+_95274707 | 3.15 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr20_+_27975549 | 2.91 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr5_-_59149625 | 2.88 |
ENSRNOT00000040641
ENSRNOT00000084216 |
Spag8
|
sperm associated antigen 8 |
| chr4_+_117780533 | 2.70 |
ENSRNOT00000021211
|
Tprkb
|
Tp53rk binding protein |
| chr4_+_45034123 | 2.66 |
ENSRNOT00000078255
|
ST7
|
suppression of tumorigenicity 7 |
| chr2_-_178521038 | 2.64 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_-_21061070 | 2.62 |
ENSRNOT00000044461
|
Olr422
|
olfactory receptor 422 |
| chr18_+_58325319 | 2.61 |
ENSRNOT00000065802
ENSRNOT00000077726 |
Napg
|
NSF attachment protein gamma |
| chr8_-_113937097 | 2.59 |
ENSRNOT00000046816
|
Nek11
|
NIMA-related kinase 11 |
| chr13_-_15395617 | 2.59 |
ENSRNOT00000046060
|
LOC304725
|
similar to contactin associated protein-like 5 isoform 1 |
| chrM_+_3904 | 2.56 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_-_199354793 | 2.56 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr7_+_18120715 | 2.52 |
ENSRNOT00000068151
|
Vom1r108
|
vomeronasal 1 receptor 108 |
| chr19_+_14459450 | 2.47 |
ENSRNOT00000079257
|
Zfp82
|
zinc finger protein 82 |
| chr3_+_67849966 | 2.47 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr4_-_9060554 | 2.36 |
ENSRNOT00000046248
|
AABR07059215.1
|
|
| chr1_-_67134827 | 2.32 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr5_+_141530211 | 2.30 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr2_-_261402880 | 2.25 |
ENSRNOT00000052396
ENSRNOT00000077839 ENSRNOT00000088561 |
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr2_+_266141581 | 2.20 |
ENSRNOT00000078187
ENSRNOT00000051951 |
Rpe65
|
RPE65, retinoid isomerohydrolase |
| chr1_-_149633029 | 2.14 |
ENSRNOT00000048861
|
LOC100910049
|
olfactory receptor 14A2-like |
| chr3_-_43117337 | 2.09 |
ENSRNOT00000078001
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr5_+_118574801 | 2.05 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr2_-_122756842 | 2.04 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chrX_-_18890090 | 2.03 |
ENSRNOT00000041039
|
Klf8
|
Kruppel-like factor 8 |
| chr4_-_121565212 | 1.93 |
ENSRNOT00000088753
|
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr20_-_46044738 | 1.80 |
ENSRNOT00000000343
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr12_-_50404550 | 1.77 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chrX_+_65800059 | 1.75 |
ENSRNOT00000076428
|
Gpr165
|
G protein-coupled receptor 165 |
| chr3_+_21114856 | 1.73 |
ENSRNOT00000044947
|
Olr423
|
olfactory receptor 423 |
| chr1_+_61268248 | 1.66 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr4_+_113782206 | 1.64 |
ENSRNOT00000009701
|
M1ap
|
meiosis 1 associated protein |
| chr1_-_67284864 | 1.62 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr7_+_23012070 | 1.49 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr2_-_88314254 | 1.47 |
ENSRNOT00000038546
|
Car13
|
carbonic anhydrase 13 |
| chr3_-_161272460 | 1.38 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr8_-_71337746 | 1.32 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr11_+_38035450 | 1.26 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr9_-_113846053 | 1.06 |
ENSRNOT00000017253
|
LOC100910483
|
ankyrin repeat domain-containing protein 12-like |
| chr1_+_64031859 | 1.03 |
ENSRNOT00000090873
|
Mboat7l1
|
membrane bound O-acyltransferase domain containing 7-like 1 |
| chr1_+_226030938 | 1.02 |
ENSRNOT00000030919
|
Fth1
|
ferritin heavy chain 1 |
| chr3_+_72080630 | 1.01 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr12_+_38368693 | 0.96 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_-_3707226 | 0.88 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr17_-_89838986 | 0.84 |
ENSRNOT00000081249
|
LOC108348078
|
ATP-dependent zinc metalloprotease YME1L1 |
| chr11_-_38420032 | 0.80 |
ENSRNOT00000002209
ENSRNOT00000080681 |
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr3_-_153454160 | 0.78 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr3_-_73401074 | 0.77 |
ENSRNOT00000012848
|
Olr476
|
olfactory receptor 476 |
| chr17_-_78561613 | 0.77 |
ENSRNOT00000020220
|
Fam107b
|
family with sequence similarity 107, member B |
| chr5_-_78183122 | 0.76 |
ENSRNOT00000002083
ENSRNOT00000067076 |
Fkbp15
|
FK506 binding protein 15 |
| chr10_+_109894286 | 0.70 |
ENSRNOT00000082209
|
Lrrc45
|
leucine rich repeat containing 45 |
| chr3_+_75712483 | 0.67 |
ENSRNOT00000049264
|
Olr575
|
olfactory receptor 575 |
| chr1_+_168546059 | 0.67 |
ENSRNOT00000086124
|
Olr101
|
olfactory receptor 101 |
| chr20_+_3363737 | 0.65 |
ENSRNOT00000079654
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr14_+_82350734 | 0.63 |
ENSRNOT00000023259
|
Tmem129
|
transmembrane protein 129 |
| chr9_-_52401660 | 0.57 |
ENSRNOT00000049387
|
Vom1r50
|
vomeronasal 1 receptor 50 |
| chr1_+_84679614 | 0.56 |
ENSRNOT00000025581
|
AABR07002774.1
|
|
| chr3_+_73115052 | 0.50 |
ENSRNOT00000090772
|
Olr454
|
olfactory receptor 454 |
| chr11_-_81379871 | 0.47 |
ENSRNOT00000089294
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr5_-_138222534 | 0.45 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chrX_-_17252877 | 0.35 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chr4_-_163445136 | 0.29 |
ENSRNOT00000080299
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr15_-_103340239 | 0.29 |
ENSRNOT00000067309
ENSRNOT00000088909 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr16_-_59721227 | 0.28 |
ENSRNOT00000015166
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr7_+_24638208 | 0.23 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr2_+_263212051 | 0.22 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr3_+_1452644 | 0.19 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr16_-_19225037 | 0.18 |
ENSRNOT00000019052
|
Klf2
|
Kruppel-like factor 2 |
| chr8_+_41247536 | 0.12 |
ENSRNOT00000073027
|
Olr1219
|
olfactory receptor 1219 |
| chr1_-_264794938 | 0.11 |
ENSRNOT00000043623
|
Pdzd7
|
PDZ domain containing 7 |
| chr9_-_97151832 | 0.11 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr8_-_77599781 | 0.03 |
ENSRNOT00000087980
|
Aqp9
|
aquaporin 9 |
| chr1_-_199655147 | 0.03 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chrX_+_23414354 | 0.02 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 32.5 | GO:0032439 | endosome localization(GO:0032439) |
| 5.2 | 15.5 | GO:0009216 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 3.5 | 14.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 3.4 | 10.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 2.8 | 11.2 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 2.4 | 9.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 2.1 | 8.4 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 2.1 | 25.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.6 | 4.7 | GO:2001201 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 1.5 | 4.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.3 | 7.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 1.1 | 3.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.1 | 3.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.1 | 37.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.0 | 7.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.9 | 8.5 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.9 | 5.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.8 | 3.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.7 | 2.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.7 | 7.8 | GO:0046549 | retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.6 | 6.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.6 | 11.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.6 | 14.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.6 | 4.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 7.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 8.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.5 | 9.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.4 | 2.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 2.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 | 5.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.4 | 5.0 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.3 | 3.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 12.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.3 | 10.6 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.3 | 13.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.3 | 1.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 4.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 3.2 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 11.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.3 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 1.6 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 4.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 0.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 5.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 6.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 13.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.2 | 1.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 9.7 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 2.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 2.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 5.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 2.6 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 3.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 11.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 8.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 12.7 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 2.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 2.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.9 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.2 | GO:0071499 | cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 3.4 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 8.0 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 1.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 11.1 | GO:0044255 | cellular lipid metabolic process(GO:0044255) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 32.5 | GO:0045179 | apical cortex(GO:0045179) |
| 2.6 | 10.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 2.3 | 11.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.1 | 37.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.5 | 4.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.4 | 5.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.1 | 13.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.8 | 11.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 3.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.6 | 7.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 4.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 5.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 8.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 8.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 2.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 7.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 9.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 1.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 13.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 5.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 3.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.0 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 1.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 5.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 32.7 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 7.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 9.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 18.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 18.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 9.1 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 3.0 | 38.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 2.6 | 10.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 2.1 | 10.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 2.1 | 14.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.6 | 7.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.1 | 12.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.0 | 10.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.9 | 8.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.9 | 2.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.8 | 8.4 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.8 | 2.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.8 | 35.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.8 | 4.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 9.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 9.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.7 | 4.7 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.6 | 1.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.5 | 11.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.5 | 9.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 13.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 5.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 3.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 2.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 0.8 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.3 | 7.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 4.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 6.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 6.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 9.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 4.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 10.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 5.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 15.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 5.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 25.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 4.0 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 5.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 6.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 5.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 11.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 25.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 6.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 11.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 10.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 10.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 10.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 5.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 4.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 9.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 9.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 32.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.4 | 6.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 7.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.3 | 12.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 4.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 10.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 2.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 9.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 8.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 4.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 10.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |