Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
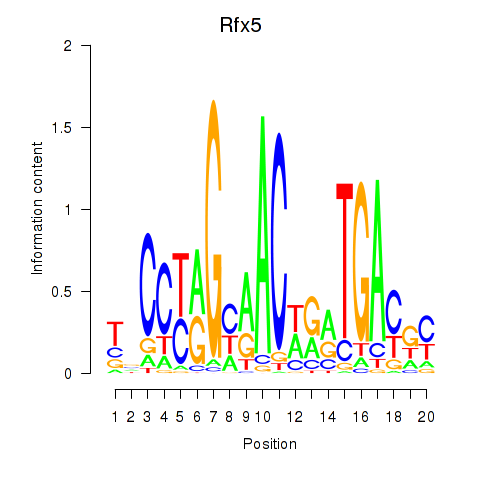
Results for Rfx5
Z-value: 1.15
Transcription factors associated with Rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx5
|
ENSRNOG00000021012 | regulatory factor X5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx5 | rn6_v1_chr2_+_196120580_196120580 | 0.34 | 2.3e-10 | Click! |
Activity profile of Rfx5 motif
Sorted Z-values of Rfx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_4043741 | 85.20 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr20_+_4087618 | 85.19 |
ENSRNOT00000000522
ENSRNOT00000060327 ENSRNOT00000080590 |
RT1-Db1
|
RT1 class II, locus Db1 |
| chr20_+_4039413 | 76.79 |
ENSRNOT00000082136
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr18_+_56071478 | 75.94 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr20_+_4020317 | 56.78 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chr20_-_4896970 | 42.69 |
ENSRNOT00000061114
ENSRNOT00000060980 |
RT1-CE7
RT1-CE5
|
RT1 class I, locus CE7 RT1 class I, locus CE5 |
| chr20_-_5234290 | 41.50 |
ENSRNOT00000073474
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr1_+_256035866 | 41.42 |
ENSRNOT00000079726
|
Kif11
|
kinesin family member 11 |
| chr20_+_3945601 | 40.18 |
ENSRNOT00000075342
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr20_-_5227620 | 40.18 |
ENSRNOT00000086240
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr20_-_157861 | 36.91 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr20_-_157665 | 36.04 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr20_+_3979035 | 32.83 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr20_+_4106189 | 32.75 |
ENSRNOT00000042571
|
RT1-Db2
|
RT1 class II, locus Db2 |
| chr20_-_5244386 | 28.28 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr20_-_3978845 | 27.69 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr20_+_5414448 | 25.92 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr13_-_72789841 | 25.81 |
ENSRNOT00000004694
|
Mr1
|
major histocompatibility complex, class I-related |
| chr20_+_3935529 | 25.59 |
ENSRNOT00000072458
|
LOC108348072
|
class II histocompatibility antigen, M alpha chain |
| chr20_+_3146856 | 24.14 |
ENSRNOT00000050159
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr20_+_5374985 | 23.73 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr20_-_1818800 | 21.32 |
ENSRNOT00000000990
|
RT1-M3-1
|
RT1 class Ib, locus M3, gene 1 |
| chr20_-_3401273 | 21.13 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr10_-_65634666 | 19.25 |
ENSRNOT00000014672
|
Foxn1
|
forkhead box N1 |
| chr3_-_7141522 | 18.03 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr20_+_3167079 | 16.62 |
ENSRNOT00000001035
|
RT1-N3
|
RT1 class Ib, locus N3 |
| chr20_-_3166569 | 16.17 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr1_+_142951094 | 13.98 |
ENSRNOT00000077441
|
Slc28a1
|
solute carrier family 28 member 1 |
| chr11_+_47243342 | 13.91 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_-_14247886 | 12.20 |
ENSRNOT00000020504
|
Nubp2
|
nucleotide binding protein 2 |
| chr1_+_101265272 | 11.89 |
ENSRNOT00000051280
|
RGD1562492
|
similar to Orphan sodium- and chloride-dependent neurotransmitter transporter NTT5 (Solute carrier family 6 member 16) |
| chr13_-_50761306 | 10.33 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr15_-_29761117 | 9.90 |
ENSRNOT00000075194
|
AABR07017658.2
|
|
| chr16_-_23975061 | 9.83 |
ENSRNOT00000090726
|
Nat1
|
N-acetyltransferase 1 |
| chr7_-_70577147 | 8.94 |
ENSRNOT00000008854
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr4_-_66380734 | 8.48 |
ENSRNOT00000009194
|
Klrg2
|
killer cell lectin like receptor G2 |
| chr1_+_189550354 | 7.91 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr1_-_170594168 | 7.84 |
ENSRNOT00000026280
|
Tpp1
|
tripeptidyl peptidase 1 |
| chr3_+_117853999 | 7.37 |
ENSRNOT00000042949
|
LOC100911847
|
40S ribosomal protein S14-like |
| chr1_-_89124132 | 6.88 |
ENSRNOT00000031044
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr12_-_17584466 | 6.68 |
ENSRNOT00000035545
|
Dnaaf5
|
dynein, axonemal, assembly factor 5 |
| chr10_-_72196437 | 6.49 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr19_-_37528011 | 6.13 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr7_+_117420788 | 5.99 |
ENSRNOT00000049198
|
Wdr97
|
WD repeat domain 97 |
| chr10_+_94466523 | 5.59 |
ENSRNOT00000014867
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr19_-_25955371 | 5.39 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr3_+_114087287 | 4.61 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr10_+_71217966 | 4.44 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr8_-_55491152 | 4.36 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr1_+_85112834 | 4.13 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr2_+_100677411 | 4.04 |
ENSRNOT00000067556
|
Ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr7_+_72985495 | 3.48 |
ENSRNOT00000008361
|
Matn2
|
matrilin 2 |
| chr10_-_15306515 | 3.13 |
ENSRNOT00000060150
|
Nhlrc4
|
NHL repeat containing 4 |
| chr4_+_181428664 | 3.10 |
ENSRNOT00000002518
|
Rep15
|
RAB15 effector protein |
| chr15_-_20032191 | 3.03 |
ENSRNOT00000032704
|
Ddhd1
|
DDHD domain containing 1 |
| chr2_-_149333597 | 2.92 |
ENSRNOT00000066948
|
P2ry14
|
purinergic receptor P2Y14 |
| chrX_+_10964067 | 2.49 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr3_-_78271875 | 2.48 |
ENSRNOT00000008475
|
Olr701
|
olfactory receptor 701 |
| chr11_-_67618704 | 1.96 |
ENSRNOT00000042208
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr1_-_22452301 | 1.59 |
ENSRNOT00000043436
|
Taar7h
|
trace amine-associated receptor 7h |
| chr14_+_73889213 | 1.09 |
ENSRNOT00000041310
ENSRNOT00000081033 |
Rab28
|
RAB28, member RAS oncogene family |
| chr13_-_71282802 | 0.82 |
ENSRNOT00000025544
|
Rgsl2h
|
regulator of G-protein signaling like 2 homolog (mouse) |
| chr1_-_89944752 | 0.32 |
ENSRNOT00000029680
|
Scgb2b24
|
secretoglobin, family 2B, member 24 |
| chr12_+_37829579 | 0.31 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 40.5 | 162.0 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 39.2 | 235.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 18.9 | 56.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 15.2 | 75.9 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 13.0 | 25.9 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 10.9 | 32.8 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 8.6 | 25.9 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 6.9 | 27.7 | GO:1901423 | response to benzene(GO:1901423) |
| 6.9 | 41.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 5.1 | 205.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 3.6 | 58.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 3.5 | 14.0 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 2.1 | 19.2 | GO:0035878 | nail development(GO:0035878) |
| 1.5 | 4.4 | GO:0061296 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.3 | 18.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.8 | 6.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.7 | 9.8 | GO:0097068 | response to thyroxine(GO:0097068) |
| 0.6 | 3.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.6 | 12.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.4 | 6.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 3.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 2.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 7.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 7.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 6.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 5.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 4.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 4.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 10.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 3.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 2.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 11.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 4.8 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.8 | 588.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 12.8 | 217.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 4.7 | 32.8 | GO:0042825 | TAP complex(GO:0042825) |
| 4.6 | 27.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.9 | 12.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 6.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.7 | 41.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.6 | 18.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.6 | 6.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 8.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 2.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 18.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 13.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 7.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 13.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 14.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.4 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.5 | 368.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 12.6 | 75.5 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 12.5 | 162.0 | GO:1990405 | protein antigen binding(GO:1990405) |
| 6.0 | 18.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 5.4 | 184.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 4.7 | 14.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 3.3 | 9.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 2.6 | 41.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 2.0 | 7.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.5 | 27.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.0 | 25.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.7 | 6.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.5 | 4.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 5.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.4 | 11.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 12.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.3 | 7.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 10.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 7.9 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 19.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 5.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 20.5 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 4.6 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 3.0 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 8.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 6.5 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 8.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 10.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 23.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 23.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 37.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.4 | 117.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.7 | 10.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 26.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.4 | 14.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 6.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 2.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 7.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |