Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
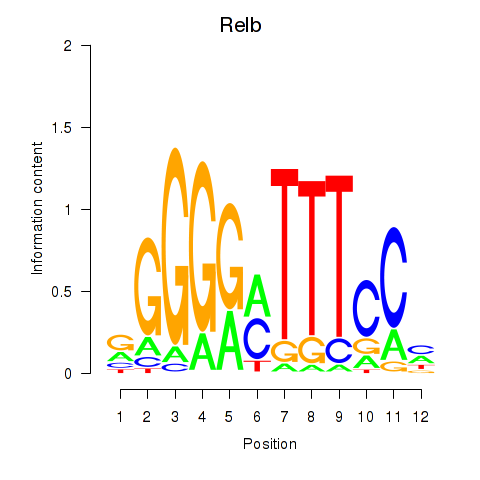
Results for Relb
Z-value: 0.62
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSRNOG00000033235 | RELB proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Relb | rn6_v1_chr1_-_80544825_80544825 | 0.59 | 1.6e-31 | Click! |
Activity profile of Relb motif
Sorted Z-values of Relb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20479999 | 26.10 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr20_-_4863011 | 25.77 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr20_-_4863198 | 25.61 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr20_+_5184515 | 24.21 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr4_+_101882994 | 23.81 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr20_-_1878126 | 22.98 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr4_+_93888502 | 20.59 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr11_+_85042348 | 13.28 |
ENSRNOT00000042220
|
AABR07034718.1
|
|
| chr6_+_109562587 | 11.92 |
ENSRNOT00000011563
|
Batf
|
basic leucine zipper ATF-like transcription factor |
| chr3_+_2643610 | 10.88 |
ENSRNOT00000037169
|
Fut7
|
fucosyltransferase 7 |
| chr13_+_47539231 | 10.70 |
ENSRNOT00000029694
|
Fcamr
|
Fc fragment of IgA and IgM receptor |
| chr1_-_80544825 | 9.24 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr1_-_89269930 | 8.84 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr17_+_56109549 | 8.48 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr5_-_136002900 | 7.98 |
ENSRNOT00000025197
|
Plk3
|
polo-like kinase 3 |
| chr5_+_168019058 | 7.93 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr20_+_4852671 | 7.69 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr3_-_105214989 | 7.03 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr20_+_4852496 | 6.67 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr4_+_87293871 | 6.30 |
ENSRNOT00000090943
|
AABR07060628.1
|
|
| chr20_-_49486550 | 5.82 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr1_-_3849080 | 4.76 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr4_-_80313411 | 4.37 |
ENSRNOT00000041621
|
RGD1565415
|
similar to ribosomal protein L27a |
| chr1_+_93242050 | 4.31 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr11_-_82366505 | 4.08 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr1_-_170579616 | 4.04 |
ENSRNOT00000076100
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr6_-_99400822 | 3.63 |
ENSRNOT00000089594
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr6_-_41039437 | 3.57 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr5_-_59025631 | 3.56 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr2_-_84437084 | 3.46 |
ENSRNOT00000077598
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr6_-_107678156 | 3.35 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr5_-_79570073 | 3.32 |
ENSRNOT00000011845
|
Tnfsf15
|
tumor necrosis factor superfamily member 15 |
| chrX_+_156655960 | 3.32 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr13_+_49870976 | 2.86 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr10_-_40296470 | 2.80 |
ENSRNOT00000086456
|
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_+_47243342 | 2.78 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr5_-_165316652 | 2.71 |
ENSRNOT00000012726
|
LOC102552055
|
angiopoietin-related protein 7-like |
| chr2_-_84436912 | 2.59 |
ENSRNOT00000059565
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr2_+_140471690 | 2.58 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr3_+_77047466 | 1.85 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chrX_-_32355296 | 1.13 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr14_+_17210733 | 0.76 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr1_-_86948845 | 0.70 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr3_+_111767161 | 0.32 |
ENSRNOT00000009430
|
Mapkbp1
|
mitogen activated protein kinase binding protein 1 |
| chr6_-_76270457 | 0.31 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr5_+_25146949 | 0.31 |
ENSRNOT00000061370
|
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr18_-_36322320 | 0.21 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Relb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 3.5 | 24.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 2.9 | 23.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.6 | 7.9 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 2.2 | 8.8 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 1.8 | 3.6 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.8 | 7.0 | GO:1900158 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.6 | 4.9 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.1 | 3.3 | GO:2000820 | positive regulation of histone H3-K9 trimethylation(GO:1900114) negative regulation of dendrite extension(GO:1903860) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.1 | 11.9 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.8 | 10.9 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.7 | 5.8 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 0.7 | 9.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.6 | 4.0 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 2.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 0.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 3.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 2.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 2.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 8.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 3.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 48.0 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 1.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 9.1 | GO:0006412 | translation(GO:0006412) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.8 | 4.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 3.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 23.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 3.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 10.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 9.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 96.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.1 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 51.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 2.1 | 10.7 | GO:0019862 | IgA binding(GO:0019862) |
| 1.8 | 10.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.4 | 7.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.2 | 23.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.0 | 41.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 3.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.4 | 7.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 2.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 8.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 3.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 2.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 8.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 20.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 9.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 11.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 14.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 7.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 9.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 54.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 7.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 5.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 8.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |