Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
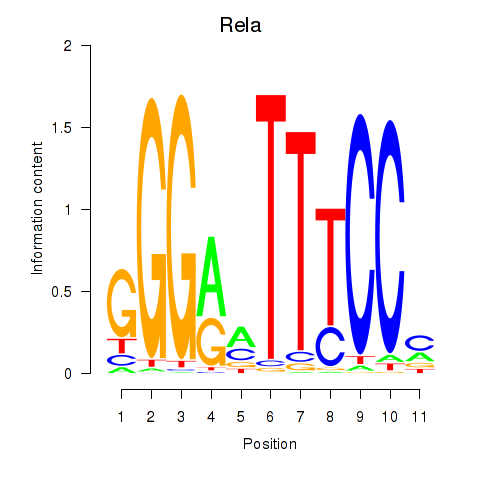
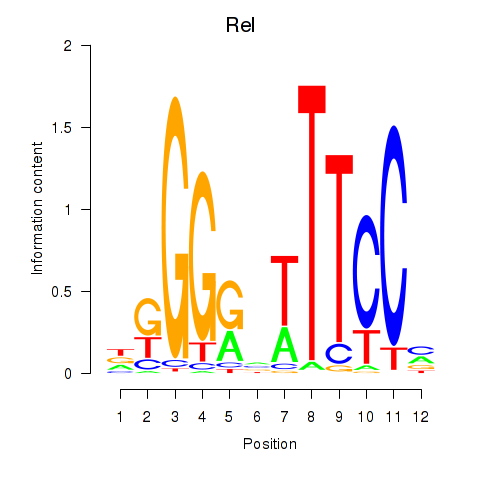
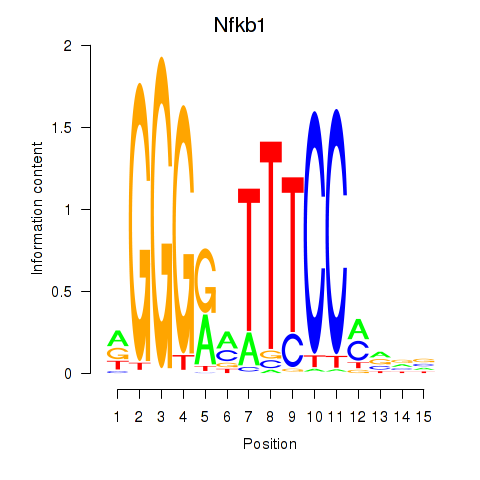
Results for Rela_Rel_Nfkb1
Z-value: 2.66
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSRNOG00000030888 | RELA proto-oncogene, NF-kB subunit |
|
Rel
|
ENSRNOG00000054437 | REL proto-oncogene, NF-kB subunit |
|
Nfkb1
|
ENSRNOG00000023258 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rel | rn6_v1_chr14_-_108509892_108509892 | 0.79 | 1.1e-70 | Click! |
| Nfkb1 | rn6_v1_chr2_-_240866689_240866689 | 0.65 | 1.5e-39 | Click! |
| Rela | rn6_v1_chr1_+_220992770_220992770 | 0.39 | 5.2e-13 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_163049084 | 214.19 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chr18_+_56071478 | 163.29 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr20_-_1878126 | 160.23 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr4_+_69384145 | 143.51 |
ENSRNOT00000084834
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr10_-_85947938 | 142.42 |
ENSRNOT00000037318
ENSRNOT00000082427 |
Arl5c
|
ADP-ribosylation factor like GTPase 5C |
| chr20_-_4863198 | 138.91 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr20_+_5184515 | 134.41 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr20_-_4863011 | 134.24 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr13_+_112031594 | 113.96 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr1_+_266053002 | 106.99 |
ENSRNOT00000026235
|
Nfkb2
|
nuclear factor kappa B subunit 2 |
| chr20_-_4070721 | 105.84 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr6_+_109562587 | 104.34 |
ENSRNOT00000011563
|
Batf
|
basic leucine zipper ATF-like transcription factor |
| chr8_-_48850671 | 96.74 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr20_+_9948908 | 94.46 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr17_+_56109549 | 89.67 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_-_80544825 | 86.13 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr10_-_70744315 | 76.64 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr2_+_189955836 | 71.87 |
ENSRNOT00000078351
|
S100a3
|
S100 calcium binding protein A3 |
| chr20_-_4921348 | 70.25 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr1_-_89269930 | 68.40 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr14_-_18745457 | 66.42 |
ENSRNOT00000003778
|
Cxcl1
|
C-X-C motif chemokine ligand 1 |
| chr6_+_135866739 | 66.03 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr8_-_6076598 | 63.83 |
ENSRNOT00000090774
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr8_+_132828091 | 62.80 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr14_-_18839420 | 60.50 |
ENSRNOT00000034090
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr1_-_7064870 | 59.71 |
ENSRNOT00000019983
|
Stx11
|
syntaxin 11 |
| chr11_+_47243342 | 59.49 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr1_-_82580761 | 59.34 |
ENSRNOT00000028131
ENSRNOT00000028132 |
Axl
|
Axl receptor tyrosine kinase |
| chr10_-_87067456 | 58.24 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr5_+_168019058 | 54.93 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr5_+_5866897 | 51.96 |
ENSRNOT00000011940
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr6_-_76270457 | 51.28 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr10_+_56187020 | 49.76 |
ENSRNOT00000046490
|
Tp53
|
tumor protein p53 |
| chr7_-_11648322 | 49.74 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr3_+_2643610 | 48.74 |
ENSRNOT00000037169
|
Fut7
|
fucosyltransferase 7 |
| chr9_+_88918433 | 48.17 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr9_-_42839837 | 45.46 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr4_+_101882994 | 44.97 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr13_+_47539231 | 44.71 |
ENSRNOT00000029694
|
Fcamr
|
Fc fragment of IgA and IgM receptor |
| chr5_-_143120165 | 42.96 |
ENSRNOT00000012314
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr3_+_177164359 | 42.59 |
ENSRNOT00000066837
|
RGD1561282
|
RGD1561282 |
| chr14_+_17195014 | 42.15 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr3_+_18315320 | 41.89 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr8_+_133029625 | 41.10 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr1_+_164502389 | 40.86 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr3_+_20375699 | 40.05 |
ENSRNOT00000088492
|
AABR07051731.1
|
|
| chrX_+_96991658 | 40.02 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr19_-_55257876 | 40.02 |
ENSRNOT00000017564
|
Cyba
|
cytochrome b-245 alpha chain |
| chr13_-_113927745 | 37.56 |
ENSRNOT00000010910
ENSRNOT00000080616 |
Cr2
|
complement C3d receptor 2 |
| chr1_+_165295847 | 37.50 |
ENSRNOT00000046943
|
P4ha3
|
prolyl 4-hydroxylase subunit alpha 3 |
| chr4_+_102876038 | 37.06 |
ENSRNOT00000085205
|
AABR07061022.3
|
|
| chr10_-_86509454 | 36.91 |
ENSRNOT00000009454
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr9_+_71230108 | 36.54 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr15_+_57241968 | 35.40 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr14_+_77067503 | 35.36 |
ENSRNOT00000085275
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr20_-_1339488 | 34.94 |
ENSRNOT00000041074
|
RT1-M2
|
RT1 class Ib, locus M2 |
| chr1_+_252589785 | 34.45 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr1_+_137014272 | 34.13 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr2_-_219097619 | 34.06 |
ENSRNOT00000078806
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr3_-_20695952 | 33.91 |
ENSRNOT00000072306
|
AABR07051746.1
|
|
| chr3_+_161519743 | 33.41 |
ENSRNOT00000055148
|
Cd40
|
CD40 molecule |
| chrX_+_134979646 | 32.59 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr13_+_92167643 | 32.53 |
ENSRNOT00000030794
|
Olr1589
|
olfactory receptor 1589 |
| chr3_+_161413298 | 32.38 |
ENSRNOT00000023965
ENSRNOT00000088776 |
Mmp9
|
matrix metallopeptidase 9 |
| chr14_+_17228856 | 32.10 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr20_+_4852671 | 31.98 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr8_+_33816386 | 31.90 |
ENSRNOT00000011925
|
Ets1
|
ETS proto-oncogene 1, transcription factor |
| chr2_-_182846061 | 31.05 |
ENSRNOT00000013025
|
Tlr2
|
toll-like receptor 2 |
| chr9_-_17835240 | 30.73 |
ENSRNOT00000026988
|
Nfkbie
|
NFKB inhibitor epsilon |
| chr10_-_13081136 | 30.40 |
ENSRNOT00000005718
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr17_+_24416651 | 30.18 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr4_+_71740532 | 30.15 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr3_-_20457554 | 29.32 |
ENSRNOT00000074237
|
AABR07051733.1
|
|
| chr3_-_20479999 | 29.16 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr4_+_157612536 | 28.69 |
ENSRNOT00000055970
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr2_+_223121410 | 28.32 |
ENSRNOT00000087559
|
AABR07013116.1
|
|
| chr17_-_10004321 | 28.27 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr4_+_93888502 | 28.27 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr20_-_157665 | 27.98 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr13_-_50916982 | 27.37 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr3_-_20419417 | 27.09 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr4_+_93791054 | 26.94 |
ENSRNOT00000042300
|
AABR07060788.1
|
|
| chr1_-_100537377 | 26.71 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr7_-_28932641 | 26.64 |
ENSRNOT00000059487
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr20_+_4852496 | 26.59 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr9_+_2190915 | 25.88 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr3_-_2853272 | 25.25 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr4_+_3043231 | 24.91 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr10_+_88764732 | 24.89 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr1_-_259357056 | 24.75 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr3_-_118959850 | 24.63 |
ENSRNOT00000092750
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr2_+_248178389 | 24.10 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr6_-_107678156 | 24.06 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr8_+_23113048 | 23.98 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr1_-_14412807 | 23.48 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr11_+_31694339 | 23.28 |
ENSRNOT00000002779
|
Ifngr2
|
interferon gamma receptor 2 |
| chr3_+_47453821 | 23.18 |
ENSRNOT00000081682
|
Tank
|
TRAF family member-associated NFKB activator |
| chr5_+_131915382 | 22.84 |
ENSRNOT00000087012
|
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr17_-_55346279 | 22.71 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr6_+_139405966 | 22.33 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr13_+_51958834 | 22.20 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr4_-_157252104 | 22.12 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr10_+_104952458 | 22.06 |
ENSRNOT00000074082
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr4_-_10517832 | 22.00 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr5_-_153924896 | 21.98 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr2_-_219458271 | 21.91 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr9_+_81427730 | 21.86 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chr17_+_80882666 | 21.78 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr10_+_104997205 | 21.77 |
ENSRNOT00000074003
|
RGD1561778
|
similar to dendritic cell-derived immunoglobulin(Ig)-like receptor 1, DIgR1 - mouse |
| chr6_+_135890931 | 21.21 |
ENSRNOT00000042973
|
Tnfaip2
|
TNF alpha induced protein 2 |
| chr3_+_120728688 | 21.18 |
ENSRNOT00000022596
|
Bcl2l11
|
BCL2 like 11 |
| chr14_-_18839595 | 21.13 |
ENSRNOT00000078746
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chrX_+_54390733 | 21.04 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr13_-_85443727 | 21.01 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr20_-_11815647 | 20.53 |
ENSRNOT00000001639
|
Itgb2
|
integrin subunit beta 2 |
| chr14_-_59735450 | 20.51 |
ENSRNOT00000071929
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_-_157155609 | 20.36 |
ENSRNOT00000016330
|
C1s
|
complement C1s |
| chr7_+_119482272 | 20.33 |
ENSRNOT00000009544
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr13_-_85443976 | 20.12 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr13_-_52088780 | 19.85 |
ENSRNOT00000008754
|
Elf3
|
E74-like factor 3 |
| chr5_-_146795866 | 19.52 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr3_+_77047466 | 19.18 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr7_+_54247460 | 18.97 |
ENSRNOT00000005361
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr20_+_4363508 | 18.92 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr2_-_157759819 | 18.77 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr10_-_40296470 | 18.58 |
ENSRNOT00000086456
|
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr9_+_42871950 | 18.56 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr7_+_116632506 | 18.54 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr20_-_49486550 | 18.50 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr8_-_132919110 | 18.43 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr14_+_17210733 | 18.37 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr20_+_3156170 | 18.35 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_-_80178708 | 18.18 |
ENSRNOT00000088676
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr1_+_221710670 | 18.13 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr11_+_67082193 | 17.91 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr3_-_125533600 | 17.90 |
ENSRNOT00000040802
|
Lrrn4
|
leucine rich repeat neuronal 4 |
| chr6_+_96479430 | 17.88 |
ENSRNOT00000006729
|
Prkch
|
protein kinase C, eta |
| chr20_+_29897594 | 17.67 |
ENSRNOT00000057752
|
Vsir
|
V-set immunoregulatory receptor |
| chr1_+_78739930 | 17.61 |
ENSRNOT00000021976
|
Strn4
|
striatin 4 |
| chr10_+_74959285 | 17.56 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr9_+_11064605 | 17.56 |
ENSRNOT00000075121
|
Stap2
|
signal transducing adaptor family member 2 |
| chr17_-_18028808 | 17.40 |
ENSRNOT00000022244
|
Kdm1b
|
lysine demethylase 1B |
| chr3_-_110324084 | 17.37 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr15_+_62406873 | 16.75 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr2_-_219090874 | 16.52 |
ENSRNOT00000019377
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chrX_+_28486869 | 16.44 |
ENSRNOT00000005620
|
Tlr7
|
toll-like receptor 7 |
| chr1_-_3849080 | 16.34 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr3_-_176958880 | 16.27 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chrX_+_156463953 | 16.25 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr1_-_87174530 | 16.00 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr4_-_23135354 | 15.82 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr3_+_80676820 | 15.75 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr6_-_41039437 | 15.68 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr10_-_83655182 | 15.50 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr6_+_139428999 | 15.49 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chr12_+_48403797 | 15.45 |
ENSRNOT00000044751
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr7_+_142912316 | 15.44 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_-_88172910 | 15.28 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr20_+_27975549 | 15.28 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr1_-_213811901 | 15.13 |
ENSRNOT00000020265
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr13_-_90814119 | 15.06 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr16_-_20148867 | 15.05 |
ENSRNOT00000040464
|
Fcho1
|
FCH domain only 1 |
| chr13_-_90977734 | 14.96 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr18_-_63488027 | 14.89 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr2_-_232245319 | 14.87 |
ENSRNOT00000014510
|
LOC691931
|
hypothetical protein LOC691931 |
| chr13_+_74456487 | 14.70 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr3_-_13978224 | 14.63 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr20_+_2057878 | 14.53 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr1_+_141120166 | 14.41 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr6_-_128727374 | 14.24 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr1_-_86948845 | 14.17 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr20_+_3979035 | 14.02 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr7_-_2353875 | 13.94 |
ENSRNOT00000074873
|
LOC100911319
|
zinc finger protein 36, C3H1 type-like 2-like |
| chr8_-_78655856 | 13.88 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr20_-_4698718 | 13.81 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr1_-_80744831 | 13.81 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr2_+_31378743 | 13.69 |
ENSRNOT00000050384
|
AABR07007853.1
|
|
| chr20_-_3299580 | 13.48 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr2_-_186333805 | 13.27 |
ENSRNOT00000022150
|
Cd1d1
|
CD1d1 molecule |
| chr4_-_103636632 | 13.22 |
ENSRNOT00000086523
|
AABR07061077.1
|
|
| chr4_-_22307453 | 13.19 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chrX_+_156429585 | 13.11 |
ENSRNOT00000083203
ENSRNOT00000077322 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr1_-_166413564 | 13.04 |
ENSRNOT00000026286
|
Atg16l2
|
autophagy related 16-like 2 |
| chr1_+_166141954 | 12.94 |
ENSRNOT00000042524
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr2_-_235852708 | 12.81 |
ENSRNOT00000009046
ENSRNOT00000005772 |
Rpl34
|
ribosomal protein L34 |
| chr10_+_34149717 | 12.75 |
ENSRNOT00000084232
ENSRNOT00000089829 |
Rack1
|
receptor for activated C kinase 1 |
| chr20_-_4390436 | 12.72 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr13_+_49870976 | 12.72 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr4_-_82141385 | 12.69 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chr2_+_185524774 | 12.67 |
ENSRNOT00000089338
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr4_+_70755795 | 12.53 |
ENSRNOT00000043527
|
LOC683849
|
similar to Anionic trypsin II precursor (Pretrypsinogen II) |
| chr1_-_82580007 | 12.49 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr5_-_76756140 | 12.48 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr5_-_29870735 | 12.47 |
ENSRNOT00000012963
|
Ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr6_-_103470427 | 12.46 |
ENSRNOT00000091560
ENSRNOT00000088795 ENSRNOT00000079824 |
Actn1
|
actinin, alpha 1 |
| chr1_+_198932870 | 12.38 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr1_-_64446818 | 12.36 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr3_+_113415774 | 12.35 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr8_+_52729003 | 12.32 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr10_-_50402616 | 12.31 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.9 | 71.8 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 32.7 | 163.3 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 31.2 | 124.7 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 24.6 | 73.9 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 20.0 | 160.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 19.9 | 59.7 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) |
| 19.5 | 234.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 19.5 | 58.6 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 19.2 | 134.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 19.2 | 76.6 | GO:0071307 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) diapedesis(GO:0050904) cellular response to vitamin K(GO:0071307) |
| 18.4 | 73.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 18.3 | 54.9 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 17.4 | 87.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 16.3 | 65.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 16.1 | 48.2 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 14.7 | 29.5 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 13.3 | 40.0 | GO:1904845 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 12.4 | 49.8 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 12.2 | 36.5 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 12.0 | 131.9 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 10.8 | 32.4 | GO:0009644 | response to high light intensity(GO:0009644) |
| 10.7 | 53.6 | GO:0030820 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) |
| 10.3 | 41.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 9.0 | 108.0 | GO:0002467 | germinal center formation(GO:0002467) |
| 8.5 | 34.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 8.0 | 31.9 | GO:0030578 | PML body organization(GO:0030578) |
| 7.9 | 7.9 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) positive regulation of dendritic cell differentiation(GO:2001200) |
| 7.5 | 15.0 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 7.3 | 73.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 7.3 | 29.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 7.3 | 21.9 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 7.1 | 28.3 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 6.8 | 20.5 | GO:0097101 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 6.8 | 115.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 6.6 | 13.3 | GO:0045402 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 6.5 | 420.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 6.4 | 90.1 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 6.4 | 25.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 6.4 | 89.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 6.3 | 63.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 6.3 | 19.0 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 6.3 | 18.9 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 6.2 | 18.7 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 6.2 | 18.5 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 6.0 | 17.9 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 5.9 | 17.7 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 5.5 | 22.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 5.4 | 21.8 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 5.4 | 5.4 | GO:0060932 | His-Purkinje system cell differentiation(GO:0060932) |
| 5.2 | 30.9 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 5.1 | 50.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 4.9 | 48.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 4.7 | 14.0 | GO:0046968 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 4.5 | 31.8 | GO:0033590 | response to cobalamin(GO:0033590) |
| 4.4 | 30.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 4.4 | 17.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 4.3 | 21.7 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 4.3 | 94.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 4.2 | 8.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 4.2 | 175.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 4.1 | 12.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 4.1 | 16.4 | GO:0045356 | toll-like receptor 7 signaling pathway(GO:0034154) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 4.1 | 28.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 4.1 | 20.5 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 4.1 | 32.6 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 4.1 | 12.2 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 3.8 | 11.5 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 3.7 | 3.7 | GO:0072683 | T cell extravasation(GO:0072683) |
| 3.6 | 28.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 3.6 | 17.9 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 3.5 | 17.3 | GO:0010159 | specification of organ position(GO:0010159) |
| 3.4 | 33.6 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 3.3 | 6.7 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 3.3 | 13.2 | GO:1905235 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) response to cyclosporin A(GO:1905237) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 3.2 | 158.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 3.1 | 15.5 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 3.0 | 48.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 3.0 | 12.0 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 3.0 | 11.8 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.9 | 8.8 | GO:1903699 | tarsal gland development(GO:1903699) |
| 2.9 | 37.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 2.8 | 42.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 2.6 | 18.5 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 2.6 | 7.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 2.6 | 25.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 2.6 | 7.7 | GO:0070268 | cornification(GO:0070268) |
| 2.6 | 15.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 2.4 | 22.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 2.4 | 7.2 | GO:1903445 | protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 2.4 | 11.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 2.3 | 16.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 2.3 | 27.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 2.2 | 11.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 2.2 | 22.0 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 2.2 | 15.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 2.2 | 24.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 2.2 | 13.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 2.1 | 6.4 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 2.1 | 2.1 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 2.1 | 8.4 | GO:1901423 | response to benzene(GO:1901423) |
| 2.1 | 16.8 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 2.1 | 35.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 2.1 | 8.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 2.1 | 10.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 2.0 | 4.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 2.0 | 15.8 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.9 | 7.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.9 | 18.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 1.9 | 5.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.9 | 7.5 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 1.8 | 9.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.8 | 14.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.8 | 14.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.8 | 12.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 1.7 | 9.9 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.6 | 8.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 1.6 | 6.4 | GO:0098501 | DNA damage response, detection of DNA damage(GO:0042769) polynucleotide dephosphorylation(GO:0098501) |
| 1.6 | 12.7 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 1.6 | 38.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 1.6 | 7.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.6 | 4.7 | GO:1905225 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) response to thyrotropin-releasing hormone(GO:1905225) |
| 1.6 | 7.8 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.5 | 6.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.5 | 21.6 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 1.5 | 19.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 1.5 | 12.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.5 | 13.5 | GO:0008228 | opsonization(GO:0008228) |
| 1.5 | 5.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.4 | 101.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 1.4 | 12.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.4 | 6.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.3 | 17.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 1.2 | 4.9 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.2 | 3.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.2 | 20.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 1.2 | 7.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 1.2 | 4.8 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.2 | 3.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 1.2 | 3.5 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.1 | 32.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 1.1 | 10.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 1.1 | 11.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 1.1 | 27.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 1.1 | 9.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 1.1 | 26.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 1.1 | 6.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.1 | 2.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.1 | 3.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 1.1 | 11.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 1.1 | 4.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.0 | 7.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.0 | 3.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 4.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.0 | 3.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.0 | 6.1 | GO:1901223 | leptin-mediated signaling pathway(GO:0033210) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.0 | 7.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 1.0 | 3.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 1.0 | 14.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 1.0 | 9.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.0 | 3.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.0 | 7.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 1.0 | 7.8 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.9 | 2.7 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.9 | 15.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.9 | 6.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.9 | 3.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 15.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.8 | 5.6 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.8 | 5.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.8 | 16.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.8 | 9.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.8 | 6.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.8 | 4.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.7 | 3.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.7 | 12.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.7 | 7.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.7 | 8.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.7 | 7.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.6 | 2.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.6 | 11.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.6 | 12.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.6 | 1.8 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.6 | 10.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.5 | 3.8 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.5 | 3.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.5 | 9.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.5 | 8.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.5 | 3.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.5 | 13.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.5 | 1.6 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.5 | 18.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.5 | 23.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.5 | 6.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.5 | 6.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.5 | 2.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.5 | 7.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.5 | 17.9 | GO:0007616 | long-term memory(GO:0007616) |
| 0.5 | 6.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.5 | 5.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.5 | 4.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.5 | 1.9 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.5 | 1.9 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.5 | 4.2 | GO:0045622 | regulation of T-helper cell differentiation(GO:0045622) |
| 0.5 | 1.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 27.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.4 | 6.9 | GO:0001553 | luteinization(GO:0001553) |
| 0.4 | 9.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.4 | 4.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.4 | 2.5 | GO:0033572 | transferrin transport(GO:0033572) iron ion import(GO:0097286) |
| 0.4 | 1.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.4 | 10.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.4 | 6.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.4 | 8.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.4 | 1.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 6.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.4 | 2.7 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.4 | 3.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.4 | 12.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.4 | 8.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.4 | 15.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 22.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.4 | 0.7 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.4 | 3.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.4 | 7.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 2.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 3.5 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.3 | 2.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.3 | 2.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 24.7 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.3 | 2.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 0.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.3 | 3.9 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.3 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 1.4 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.3 | 4.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.3 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 2.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 14.9 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.2 | 1.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 10.9 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.2 | 23.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 7.3 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 3.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 3.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 0.7 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 1.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 1.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 6.9 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.2 | 2.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 1.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 3.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 2.0 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 2.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.2 | GO:0033233 | regulation of protein sumoylation(GO:0033233) negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 1.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.5 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 1.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 7.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.2 | 10.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 3.6 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.6 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 3.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 12.5 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 2.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 1.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.4 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 77.0 | GO:0006955 | immune response(GO:0006955) |
| 0.1 | 3.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 2.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 2.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.4 | GO:0034627 | tryptophan catabolic process to kynurenine(GO:0019441) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 5.8 | GO:0031334 | positive regulation of protein complex assembly(GO:0031334) |
| 0.1 | 0.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 1.0 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.1 | 0.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 9.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 3.6 | GO:0048754 | branching morphogenesis of an epithelial tube(GO:0048754) |
| 0.0 | 4.0 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 1.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 2.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 5.1 | GO:1901653 | cellular response to peptide(GO:1901653) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 5.5 | GO:0006412 | translation(GO:0006412) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 54.4 | 163.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 30.3 | 272.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 12.7 | 114.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 12.6 | 50.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 12.2 | 36.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 10.3 | 31.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 9.4 | 37.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 8.3 | 24.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 7.1 | 105.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 6.9 | 34.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 6.1 | 98.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 6.1 | 43.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 5.5 | 71.8 | GO:0033643 | host cell part(GO:0033643) |
| 5.1 | 20.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 5.0 | 50.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 4.7 | 14.0 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 4.4 | 21.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 4.3 | 21.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 4.1 | 12.2 | GO:0060187 | cell pole(GO:0060187) |
| 3.7 | 92.4 | GO:0000145 | exocyst(GO:0000145) |
| 3.7 | 25.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 3.2 | 12.7 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 3.1 | 40.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 3.0 | 35.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 2.8 | 8.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 2.8 | 137.0 | GO:0016235 | aggresome(GO:0016235) |
| 2.8 | 22.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.3 | 13.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.1 | 25.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.0 | 14.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 2.0 | 12.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 2.0 | 618.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 1.9 | 11.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.8 | 44.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.8 | 9.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 1.7 | 8.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 1.7 | 64.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 1.6 | 16.4 | GO:0032009 | early phagosome(GO:0032009) |
| 1.6 | 7.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.6 | 7.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.3 | 6.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.3 | 3.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.2 | 28.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 1.2 | 62.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 1.2 | 14.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.2 | 16.8 | GO:0042581 | specific granule(GO:0042581) |
| 1.1 | 3.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.1 | 20.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 1.0 | 7.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.0 | 21.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.9 | 2.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 3.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.9 | 6.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.8 | 53.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.8 | 3.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.7 | 64.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.7 | 3.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.7 | 46.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.7 | 66.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.7 | 3.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 3.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.7 | 17.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 11.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.6 | 12.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.6 | 11.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 12.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 26.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 3.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 54.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.5 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 671.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.5 | 6.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 1.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.5 | 3.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 2.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 15.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.4 | 5.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.3 | GO:0036396 | MIS complex(GO:0036396) |
| 0.4 | 2.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 7.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.3 | 10.4 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 3.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.3 | 3.7 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 103.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.3 | 2.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 7.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 9.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 1.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 5.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 15.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.3 | 5.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 15.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 2.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 4.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 29.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 17.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 67.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 25.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 3.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 8.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 66.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.2 | 1.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 2.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 53.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 5.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 6.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 4.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 9.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 7.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 6.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 202.8 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.1 | 2.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 7.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 8.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 11.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 3.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 9.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 4.5 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 61.5 | 307.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 27.2 | 163.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 18.5 | 92.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 18.1 | 163.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 14.4 | 129.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 13.6 | 40.9 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) vasopressin receptor binding(GO:0031893) |
| 13.3 | 13.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 12.8 | 76.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 12.2 | 36.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 12.0 | 48.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 11.7 | 164.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 10.3 | 31.0 | GO:0042498 | diacyl lipopeptide binding(GO:0042498) |
| 10.3 | 30.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 9.4 | 37.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 9.3 | 166.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 8.9 | 44.7 | GO:0019862 | IgA binding(GO:0019862) |
| 8.5 | 34.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 8.3 | 41.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 8.1 | 48.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 7.4 | 29.8 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 6.2 | 49.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 6.2 | 68.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 5.4 | 16.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 5.1 | 212.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 5.0 | 14.9 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 4.7 | 23.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 4.6 | 88.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 4.6 | 50.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 4.5 | 50.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 4.5 | 17.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 4.4 | 21.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 4.3 | 8.6 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 4.2 | 173.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 4.2 | 24.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 4.1 | 12.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 3.9 | 23.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 3.9 | 101.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 3.8 | 18.9 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 3.7 | 41.2 | GO:0019864 | IgG binding(GO:0019864) |
| 3.7 | 11.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 3.5 | 35.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 3.5 | 28.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 3.4 | 37.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 3.3 | 29.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 3.1 | 18.5 | GO:0035473 | lipase binding(GO:0035473) |
| 2.9 | 17.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 2.9 | 11.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 2.7 | 8.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 2.6 | 13.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 2.6 | 15.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.6 | 10.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 2.6 | 7.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.6 | 7.9 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 2.6 | 68.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 2.5 | 20.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.5 | 22.1 | GO:0046703 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) natural killer cell lectin-like receptor binding(GO:0046703) |
| 2.4 | 19.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 2.3 | 16.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 2.3 | 4.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 2.3 | 11.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 2.2 | 9.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 2.2 | 8.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 2.1 | 6.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 2.1 | 6.4 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 2.1 | 12.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 2.0 | 12.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 2.0 | 17.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.8 | 75.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.8 | 12.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.8 | 10.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 1.7 | 8.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.6 | 4.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.6 | 27.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 1.6 | 8.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.6 | 12.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.6 | 12.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.5 | 49.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 1.5 | 6.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.5 | 15.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 1.5 | 6.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.5 | 4.5 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.4 | 59.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 1.4 | 5.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.3 | 39.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 1.3 | 9.0 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 1.3 | 3.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.3 | 3.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.2 | 31.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.2 | 3.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.2 | 7.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 1.2 | 25.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.2 | 3.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.1 | 5.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 1.1 | 7.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.1 | 8.5 | GO:0031386 | protein tag(GO:0031386) |
| 1.0 | 15.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 1.0 | 3.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.0 | 5.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.0 | 8.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.9 | 9.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.9 | 16.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.9 | 21.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.9 | 8.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.9 | 6.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.9 | 275.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.9 | 3.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 44.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.8 | 5.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.8 | 29.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.8 | 3.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.8 | 19.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.8 | 7.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.8 | 1.5 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.8 | 8.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 13.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.7 | 229.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.7 | 5.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.7 | 7.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.7 | 13.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.7 | 6.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.7 | 6.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.6 | 17.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 10.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 12.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.6 | 1.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.6 | 1.7 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.6 | 16.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 4.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.5 | 50.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.5 | 6.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.5 | 6.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 61.5 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.5 | 24.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.5 | 5.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.5 | 4.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 21.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 1.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 4.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 2.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.4 | 3.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 0.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 7.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 8.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 3.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 4.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 59.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 2.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 7.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.6 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 5.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.3 | 3.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 2.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 17.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 2.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 8.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 11.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 33.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.3 | 4.6 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.3 | 1.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 1.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.3 | 5.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 66.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 17.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 8.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 4.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 12.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 7.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 3.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 49.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 3.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 25.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 13.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.2 | 1.1 | GO:0043995 | G-quadruplex RNA binding(GO:0002151) histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 2.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 3.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 2.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 76.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 7.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 11.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.2 | 42.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 5.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 8.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 7.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 7.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 11.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 2.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 19.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 6.6 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 2.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.9 | GO:0022824 | transmitter-gated ion channel activity(GO:0022824) transmitter-gated channel activity(GO:0022835) |
| 0.1 | 19.5 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) |
| 0.1 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 7.5 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 2.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 291.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 8.4 | 92.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 6.9 | 13.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 6.7 | 114.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 4.2 | 58.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 3.8 | 34.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 3.7 | 26.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 3.7 | 148.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 3.6 | 132.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 3.3 | 36.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 3.3 | 82.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 3.3 | 141.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 3.2 | 100.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 3.1 | 82.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 2.7 | 54.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 2.7 | 62.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 2.4 | 48.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 2.3 | 36.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 2.0 | 43.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.9 | 21.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.9 | 36.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 1.9 | 70.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 1.9 | 7.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 1.8 | 37.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 1.5 | 16.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.5 | 79.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.4 | 409.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 1.4 | 21.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 1.3 | 43.7 | PID FGF PATHWAY | FGF signaling pathway |
| 1.2 | 17.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.1 | 19.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 1.0 | 33.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 1.0 | 14.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 1.0 | 10.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 1.0 | 41.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.9 | 18.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.9 | 20.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.8 | 29.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.8 | 9.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.8 | 22.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.7 | 28.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.7 | 19.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.6 | 61.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.6 | 6.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.5 | 26.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.5 | 7.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 20.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.5 | 6.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 17.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.4 | 3.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 7.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.4 | 8.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 6.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 3.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 5.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 6.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 5.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 5.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 4.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 15.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 31.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 13.7 | 686.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 11.3 | 191.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 4.2 | 24.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 4.0 | 88.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 3.8 | 107.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 3.5 | 58.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 3.4 | 40.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 3.3 | 60.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 3.1 | 59.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 2.9 | 34.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 2.8 | 96.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 2.8 | 8.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 2.7 | 16.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 2.6 | 52.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 2.5 | 24.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.3 | 114.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 2.1 | 35.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 2.0 | 28.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 2.0 | 14.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.9 | 20.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.7 | 26.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.7 | 20.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 1.5 | 41.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.5 | 25.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.5 | 130.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 1.4 | 25.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 1.4 | 49.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 1.2 | 16.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.2 | 32.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.1 | 14.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 1.1 | 11.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 1.0 | 81.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 1.0 | 16.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.9 | 25.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.8 | 45.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.8 | 24.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.8 | 48.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.8 | 24.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.7 | 16.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 2.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 13.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.6 | 8.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.6 | 8.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 9.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.5 | 3.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.5 | 2.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 4.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 41.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 7.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 60.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.3 | 3.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 3.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 4.6 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.3 | 4.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 7.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 6.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 13.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 4.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 13.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 7.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 3.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 9.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 3.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 2.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 3.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |