Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
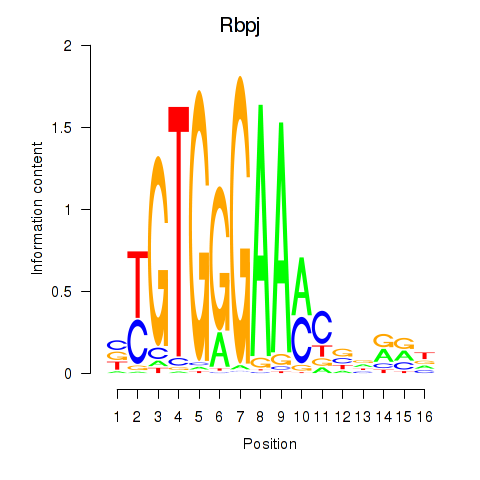
Results for Rbpj
Z-value: 0.72
Transcription factors associated with Rbpj
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rbpj
|
ENSRNOG00000007797 | recombination signal binding protein for immunoglobulin kappa J region |
|
Rbpj
|
ENSRNOG00000046327 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rbpj | rn6_v1_chr14_-_59735450_59735450 | -0.12 | 3.2e-02 | Click! |
Activity profile of Rbpj motif
Sorted Z-values of Rbpj motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_55822551 | 26.46 |
ENSRNOT00000082624
|
Lrp2
|
LDL receptor related protein 2 |
| chr13_+_56598957 | 22.98 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr5_-_78985990 | 20.22 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr10_+_71159869 | 18.85 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr10_+_56662561 | 18.83 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr16_-_81797815 | 18.03 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr1_+_191829555 | 18.01 |
ENSRNOT00000067138
|
Scnn1b
|
sodium channel epithelial 1 beta subunit |
| chr4_-_154855098 | 16.61 |
ENSRNOT00000041957
|
LOC297568
|
alpha-1-inhibitor III |
| chr4_+_57952982 | 15.37 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chr3_-_161212188 | 15.28 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr10_+_56662242 | 15.02 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr2_-_236480502 | 15.01 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr7_+_141237768 | 14.07 |
ENSRNOT00000091717
|
Aqp2
|
aquaporin 2 |
| chr1_-_189181901 | 12.34 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr3_-_101465995 | 11.12 |
ENSRNOT00000080175
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr10_-_88036040 | 10.16 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr7_+_116745061 | 9.27 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr2_-_113616766 | 9.26 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chrX_-_139079336 | 9.18 |
ENSRNOT00000047605
|
Usp26
|
ubiquitin specific peptidase 26 |
| chr1_-_85220237 | 8.98 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr14_+_34727915 | 8.97 |
ENSRNOT00000085089
|
Kdr
|
kinase insert domain receptor |
| chr14_+_34727623 | 8.67 |
ENSRNOT00000071405
ENSRNOT00000090183 |
Kdr
|
kinase insert domain receptor |
| chr8_+_69971778 | 8.27 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr1_-_226152524 | 7.94 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chrX_+_68891227 | 7.81 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chr8_+_119160731 | 7.44 |
ENSRNOT00000046745
|
Als2cl
|
ALS2 C-terminal like |
| chr11_-_74315248 | 7.31 |
ENSRNOT00000002346
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr17_-_55709740 | 7.03 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr2_-_233743866 | 6.91 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr3_+_45683993 | 6.76 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr9_+_119517101 | 6.67 |
ENSRNOT00000020476
|
Lpin2
|
lipin 2 |
| chrX_-_112473822 | 6.61 |
ENSRNOT00000079180
|
Col4a6
|
collagen type IV alpha 6 chain |
| chr11_+_81972219 | 6.47 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr16_-_74864816 | 6.40 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chrX_+_110894845 | 6.27 |
ENSRNOT00000080072
|
Tbc1d8b
|
TBC1 domain family member 8B |
| chr11_-_14160697 | 6.18 |
ENSRNOT00000081096
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr16_+_49485256 | 6.17 |
ENSRNOT00000059317
|
RGD1564308
|
similar to LOC495042 protein |
| chr12_-_23661009 | 5.97 |
ENSRNOT00000059451
|
Upk3bl
|
uroplakin 3B-like |
| chr8_-_55171718 | 5.53 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr2_-_178334900 | 5.45 |
ENSRNOT00000090152
|
Fnip2
|
folliculin interacting protein 2 |
| chr4_+_109497962 | 5.41 |
ENSRNOT00000057869
|
Reg1a
|
regenerating family member 1 alpha |
| chr18_+_56379890 | 5.12 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr19_+_43163129 | 5.09 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr18_-_31071371 | 4.96 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr11_-_14160892 | 4.82 |
ENSRNOT00000091864
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr12_-_47046022 | 4.80 |
ENSRNOT00000001539
|
Srsf9
|
serine and arginine rich splicing factor 9 |
| chr10_-_55783489 | 4.54 |
ENSRNOT00000010415
|
Alox15b
|
arachidonate 15-lipoxygenase, type B |
| chr5_-_24255606 | 4.50 |
ENSRNOT00000037483
|
Plekhf2
|
pleckstrin homology and FYVE domain containing 2 |
| chr19_+_43163562 | 4.49 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_+_64506735 | 4.46 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chrX_-_73778595 | 4.33 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr7_+_117055395 | 4.32 |
ENSRNOT00000091960
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr6_+_34094306 | 4.24 |
ENSRNOT00000051970
|
Wdr35
|
WD repeat domain 35 |
| chr16_-_19777414 | 3.86 |
ENSRNOT00000022898
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr1_-_216274930 | 3.85 |
ENSRNOT00000040850
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr10_-_85636902 | 3.79 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr1_+_101213788 | 3.79 |
ENSRNOT00000090838
|
Tead2
|
TEA domain transcription factor 2 |
| chr6_-_88917070 | 3.77 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chrX_-_77061604 | 3.77 |
ENSRNOT00000083643
ENSRNOT00000089386 |
Magt1
|
magnesium transporter 1 |
| chr18_-_37245809 | 3.70 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_+_82587420 | 3.70 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr16_+_78539489 | 3.53 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr1_+_216254979 | 3.50 |
ENSRNOT00000046669
|
Tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr1_+_101214593 | 3.48 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr19_+_37282018 | 3.36 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr9_+_81880177 | 3.32 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr17_-_45154355 | 3.29 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr7_+_117055623 | 3.27 |
ENSRNOT00000012461
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr6_+_107169528 | 3.23 |
ENSRNOT00000012495
|
Psen1
|
presenilin 1 |
| chr1_+_162342051 | 3.20 |
ENSRNOT00000016478
|
Alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr1_+_185427736 | 3.17 |
ENSRNOT00000064706
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr4_-_30380119 | 3.08 |
ENSRNOT00000036460
|
Pon2
|
paraoxonase 2 |
| chr1_-_100671074 | 2.95 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr16_-_23991570 | 2.68 |
ENSRNOT00000044198
ENSRNOT00000018854 |
Nat2
Nat1
|
N-acetyltransferase 2 N-acetyltransferase 1 |
| chr5_+_146383942 | 2.55 |
ENSRNOT00000078588
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr7_-_92882068 | 2.38 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chrX_+_112020646 | 2.37 |
ENSRNOT00000079841
|
Mid2
|
midline 2 |
| chr1_+_252894663 | 2.36 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr6_+_111534548 | 2.29 |
ENSRNOT00000087462
|
AABR07065139.1
|
|
| chr17_-_22678253 | 2.24 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr2_+_86951776 | 2.21 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
| chr12_-_38782010 | 2.20 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr19_+_61334168 | 2.09 |
ENSRNOT00000080465
|
Nrp1
|
neuropilin 1 |
| chr1_+_277190964 | 2.07 |
ENSRNOT00000080511
|
Casp7
|
caspase 7 |
| chr7_+_11582984 | 2.04 |
ENSRNOT00000026893
|
Gng7
|
G protein subunit gamma 7 |
| chr7_+_11033317 | 2.01 |
ENSRNOT00000007498
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr10_-_17075139 | 1.96 |
ENSRNOT00000039398
|
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr1_+_199412834 | 1.95 |
ENSRNOT00000031891
|
Fus
|
fused in sarcoma RNA binding protein |
| chr1_+_157573324 | 1.89 |
ENSRNOT00000092066
|
Rab30
|
RAB30, member RAS oncogene family |
| chr19_-_37281933 | 1.89 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr3_+_7279340 | 1.84 |
ENSRNOT00000017029
|
Ak8
|
adenylate kinase 8 |
| chr8_-_104912959 | 1.80 |
ENSRNOT00000017363
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr12_+_19314251 | 1.79 |
ENSRNOT00000001827
|
Ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr1_+_79931156 | 1.52 |
ENSRNOT00000019814
ENSRNOT00000084985 |
Sympk
|
symplekin |
| chr8_+_61659445 | 1.42 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr1_+_261180007 | 1.41 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr13_+_110257571 | 1.41 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr16_-_49484985 | 1.38 |
ENSRNOT00000016723
|
Ufsp2
|
UFM1-specific peptidase 2 |
| chr2_-_188587498 | 1.28 |
ENSRNOT00000027878
|
Slc50a1
|
solute carrier family 50 member 1 |
| chr3_-_3834186 | 1.27 |
ENSRNOT00000061669
|
Sdccag3
|
serologically defined colon cancer antigen 3 |
| chr14_+_4125380 | 1.23 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr6_-_103470427 | 1.13 |
ENSRNOT00000091560
ENSRNOT00000088795 ENSRNOT00000079824 |
Actn1
|
actinin, alpha 1 |
| chr2_-_181102918 | 1.12 |
ENSRNOT00000017190
|
Gucy1a3
|
guanylate cyclase 1 soluble subunit alpha 3 |
| chr18_+_58325319 | 0.99 |
ENSRNOT00000065802
ENSRNOT00000077726 |
Napg
|
NSF attachment protein gamma |
| chr3_+_11587941 | 0.94 |
ENSRNOT00000071505
|
Dpm2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chrX_-_105417323 | 0.93 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr6_-_124177404 | 0.92 |
ENSRNOT00000005369
|
Nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr5_+_154800226 | 0.91 |
ENSRNOT00000016046
|
Htr1d
|
5-hydroxytryptamine receptor 1D |
| chr16_+_8350326 | 0.87 |
ENSRNOT00000027071
|
Parg
|
poly (ADP-ribose) glycohydrolase |
| chr6_-_67085390 | 0.86 |
ENSRNOT00000083277
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr11_-_87628502 | 0.84 |
ENSRNOT00000002568
|
Med15
|
mediator complex subunit 15 |
| chrX_+_128493614 | 0.75 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr1_-_226630900 | 0.62 |
ENSRNOT00000028092
|
Tmem138
|
transmembrane protein 138 |
| chr12_-_29919320 | 0.53 |
ENSRNOT00000038092
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog |
| chr10_+_55707164 | 0.44 |
ENSRNOT00000009757
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr10_-_15346236 | 0.38 |
ENSRNOT00000088104
ENSRNOT00000027430 |
Capn15
|
calpain 15 |
| chr3_+_161212156 | 0.32 |
ENSRNOT00000020280
ENSRNOT00000083553 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr2_+_184600721 | 0.30 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr12_+_19328957 | 0.29 |
ENSRNOT00000033288
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr15_-_60766579 | 0.29 |
ENSRNOT00000079978
|
Akap11
|
A-kinase anchoring protein 11 |
| chr1_+_87697005 | 0.28 |
ENSRNOT00000028158
|
Zfp84
|
zinc finger protein 84 |
| chr8_+_117170620 | 0.26 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr7_-_145174771 | 0.25 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr20_-_6500523 | 0.23 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr17_+_76532611 | 0.20 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr7_+_123043503 | 0.16 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr3_+_37599728 | 0.15 |
ENSRNOT00000090575
|
Rif1
|
replication timing regulatory factor 1 |
| chrX_-_112328642 | 0.08 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr20_+_33174892 | 0.06 |
ENSRNOT00000045139
|
LOC100361475
|
transforming growth factor beta regulator 1-like |
| chr5_+_73496027 | 0.05 |
ENSRNOT00000022395
|
Actl7a
|
actin-like 7a |
| chr9_-_81880105 | 0.02 |
ENSRNOT00000022677
|
Rnf25
|
ring finger protein 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rbpj
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0061296 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 4.7 | 14.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 3.1 | 12.3 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.9 | 17.6 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 2.4 | 7.3 | GO:0021558 | trochlear nerve development(GO:0021558) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 2.1 | 15.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 2.0 | 20.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.9 | 26.5 | GO:0045056 | transcytosis(GO:0045056) |
| 1.9 | 11.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.8 | 3.7 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.8 | 5.4 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 1.5 | 4.5 | GO:0045751 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 1.4 | 4.3 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 1.4 | 6.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.3 | 5.1 | GO:0072277 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.1 | 3.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.0 | 2.1 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.9 | 4.5 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.8 | 10.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.8 | 2.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.7 | 9.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.7 | 7.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 3.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 3.2 | GO:1904796 | synaptic vesicle targeting(GO:0016080) regulation of core promoter binding(GO:1904796) |
| 0.5 | 18.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.5 | 2.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.5 | 7.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.4 | 4.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.4 | 2.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.4 | 7.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.4 | 1.1 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.4 | 7.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.4 | 18.0 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 5.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 6.8 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.3 | 8.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.3 | 7.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.3 | 2.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 3.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 3.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 2.7 | GO:0097068 | response to thyroxine(GO:0097068) |
| 0.3 | 1.3 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.2 | 0.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 3.9 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 7.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 2.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 4.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 0.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 2.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 3.5 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 4.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 33.9 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 31.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 16.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 9.0 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 | 0.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.9 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 | 9.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.9 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 1.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 3.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 5.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 1.4 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 3.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 2.4 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 1.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 1.9 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0021510 | spinal cord development(GO:0021510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 2.5 | 19.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.4 | 7.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.8 | 6.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.8 | 18.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.7 | 3.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 2.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 4.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 3.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.5 | 3.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 6.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 5.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 14.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 4.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 25.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.3 | 0.9 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 3.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 2.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 1.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 20.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 16.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 10.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 9.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 39.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 5.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 7.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 41.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 5.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 26.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 4.0 | 20.2 | GO:0019862 | IgA binding(GO:0019862) |
| 3.1 | 24.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 3.0 | 15.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 2.8 | 14.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 2.1 | 6.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 2.0 | 18.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 2.0 | 7.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 1.1 | 3.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.0 | 3.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.9 | 3.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.9 | 2.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.8 | 9.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 5.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.7 | 2.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.6 | 7.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.6 | 15.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 3.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 1.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.4 | 33.9 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.4 | 6.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 7.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 1.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 3.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 0.9 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 6.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 4.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 7.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 3.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 3.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 11.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.2 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 7.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 4.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 7.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 16.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 15.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 12.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 14.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.8 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 6.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 10.6 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.1 | 1.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 13.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 3.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 5.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 1.3 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.1 | 26.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 5.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 18.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 11.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 7.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 2.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 9.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 3.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 7.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 30.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 9.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 8.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 23.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.8 | 18.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.6 | 19.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.1 | 13.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 26.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.8 | 7.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.7 | 25.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.5 | 6.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 15.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 3.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 10.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 3.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.3 | 7.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 6.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 11.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 2.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |