Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Rarb
Z-value: 0.43
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSRNOG00000024061 | retinoic acid receptor, beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarb | rn6_v1_chr15_+_10120206_10120206 | 0.49 | 1.7e-20 | Click! |
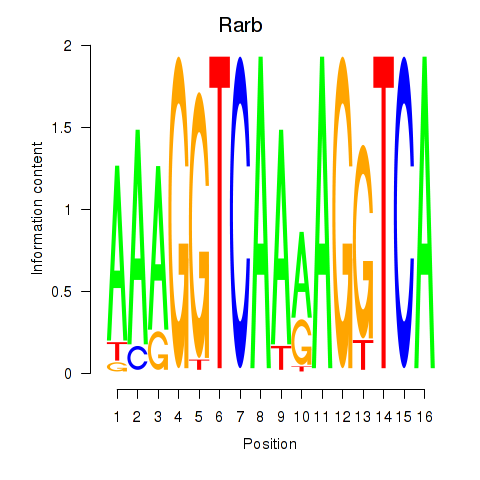
Activity profile of Rarb motif
Sorted Z-values of Rarb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_48360131 | 25.03 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr14_+_22375955 | 17.97 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr6_+_33176778 | 13.26 |
ENSRNOT00000046811
ENSRNOT00000007371 |
Apob
|
apolipoprotein B |
| chr4_-_82203154 | 12.43 |
ENSRNOT00000086210
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr16_-_20460959 | 10.88 |
ENSRNOT00000026457
|
Pde4c
|
phosphodiesterase 4C |
| chr1_+_264741911 | 10.20 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr18_+_61261418 | 8.64 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr14_+_22806132 | 7.90 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr17_-_78499881 | 4.37 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr9_+_30419001 | 4.28 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chr14_+_22597103 | 3.53 |
ENSRNOT00000048482
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr10_+_70627401 | 1.78 |
ENSRNOT00000076897
|
Rasl10b
|
RAS-like, family 10, member B |
| chr9_+_97355924 | 1.68 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr19_-_55389256 | 1.15 |
ENSRNOT00000064180
|
Aprt
|
adenine phosphoribosyl transferase |
| chr2_-_250923744 | 0.94 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr1_-_22570303 | 0.73 |
ENSRNOT00000035539
|
Taar3
|
trace amine-associated receptor 3 |
| chr18_+_30885789 | 0.62 |
ENSRNOT00000044434
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr8_+_40488792 | 0.30 |
ENSRNOT00000076868
|
Olr1204
|
olfactory receptor 1204 |
| chr1_-_245817809 | 0.27 |
ENSRNOT00000080607
|
AABR07006672.1
|
|
| chr10_+_55555089 | 0.19 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr13_+_110511668 | 0.16 |
ENSRNOT00000006235
|
Nek2
|
NIMA-related kinase 2 |
| chr1_+_247110216 | 0.12 |
ENSRNOT00000016000
|
Cdc37l1
|
cell division cycle 37-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 25.0 | GO:0015871 | choline transport(GO:0015871) |
| 1.9 | 13.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 1.8 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.4 | 10.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.4 | 1.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 3.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 4.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 1.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 4.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.4 | 4.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 25.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 18.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 10.9 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 25.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 2.2 | 13.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.9 | 29.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 10.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 1.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.2 | 1.7 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 12.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 10.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 25.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.8 | 13.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 11.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 10.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |