Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
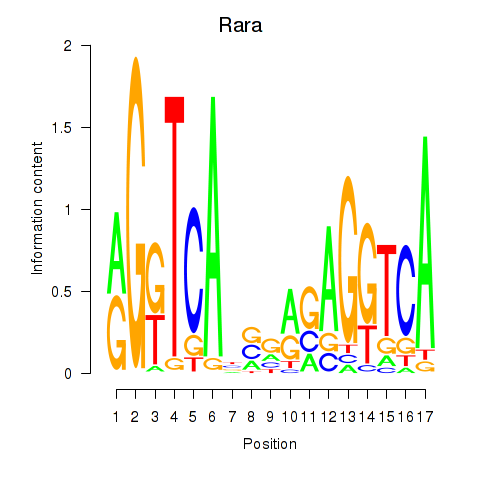
Results for Rara
Z-value: 0.74
Transcription factors associated with Rara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rara
|
ENSRNOG00000009972 | retinoic acid receptor, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rara | rn6_v1_chr10_+_86860685_86860685 | 0.32 | 3.3e-09 | Click! |
Activity profile of Rara motif
Sorted Z-values of Rara motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_149624712 | 26.91 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr2_+_188543137 | 25.82 |
ENSRNOT00000027850
|
Muc1
|
mucin 1, cell surface associated |
| chrX_+_139780386 | 20.95 |
ENSRNOT00000081147
|
AABR07041779.2
|
|
| chr4_-_17594598 | 20.91 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr5_-_50193571 | 19.17 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr4_-_11610518 | 18.79 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr13_+_47454591 | 17.09 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr10_+_90348299 | 14.01 |
ENSRNOT00000089616
|
Rundc3a
|
RUN domain containing 3A |
| chr4_+_96562725 | 13.73 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr10_-_82229140 | 12.52 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr1_-_173682226 | 11.94 |
ENSRNOT00000020137
|
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr7_+_30699476 | 11.77 |
ENSRNOT00000059269
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_-_14056169 | 11.62 |
ENSRNOT00000017833
|
Syngr3
|
synaptogyrin 3 |
| chr8_-_107656861 | 10.96 |
ENSRNOT00000050374
|
Mras
|
muscle RAS oncogene homolog |
| chr13_-_52197205 | 9.99 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr1_-_64350338 | 9.95 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr20_-_31956649 | 9.87 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr1_-_213973163 | 8.98 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chrX_+_105537602 | 8.66 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr5_-_141242131 | 8.13 |
ENSRNOT00000081482
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_123193130 | 8.05 |
ENSRNOT00000019554
|
Anxa5
|
annexin A5 |
| chr18_+_60639860 | 7.87 |
ENSRNOT00000083363
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_-_152473868 | 7.54 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr19_+_10563423 | 7.51 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr16_+_21061237 | 7.38 |
ENSRNOT00000092136
|
Ncan
|
neurocan |
| chr18_+_48434652 | 7.07 |
ENSRNOT00000047755
|
Prdm6
|
PR/SET domain 6 |
| chr12_-_40590361 | 6.74 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr1_+_100299626 | 6.43 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr8_-_65587658 | 6.38 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr6_+_111049559 | 6.24 |
ENSRNOT00000015571
|
Tmem63c
|
transmembrane protein 63c |
| chr16_+_20432899 | 6.11 |
ENSRNOT00000026271
|
Mpv17l2
|
MPV17 mitochondrial inner membrane protein like 2 |
| chr7_+_130532435 | 5.06 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr15_-_56970365 | 5.04 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr3_+_20641664 | 4.89 |
ENSRNOT00000044699
|
AABR07051741.1
|
|
| chr10_+_57218087 | 4.59 |
ENSRNOT00000089853
|
Mink1
|
misshapen-like kinase 1 |
| chr18_-_86878142 | 4.39 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chr2_+_178679395 | 4.37 |
ENSRNOT00000088686
|
Fam198b
|
family with sequence similarity 198, member B |
| chr7_-_139223116 | 4.16 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr8_-_47393503 | 4.09 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr1_+_221538104 | 3.88 |
ENSRNOT00000028527
|
Batf2
|
basic leucine zipper ATF-like transcription factor 2 |
| chr8_-_127900463 | 3.69 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr15_-_4057104 | 3.65 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr11_-_64968437 | 3.38 |
ENSRNOT00000059541
|
Cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr14_+_85230648 | 3.11 |
ENSRNOT00000089866
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chrX_+_96991658 | 2.90 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr4_+_101882994 | 2.88 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr2_+_178679041 | 2.85 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr8_-_50277797 | 2.60 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr3_-_20518105 | 2.53 |
ENSRNOT00000050435
|
AABR07051735.1
|
|
| chr12_+_50270189 | 2.52 |
ENSRNOT00000086782
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr3_-_20419417 | 2.37 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr20_+_42966140 | 2.05 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr10_-_56444847 | 2.03 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr2_+_62887267 | 2.02 |
ENSRNOT00000048026
|
Drosha
|
drosha ribonuclease III |
| chr1_-_227486976 | 1.77 |
ENSRNOT00000035812
|
Ms4a14
|
membrane spanning 4-domains A14 |
| chr10_+_68232094 | 1.68 |
ENSRNOT00000009172
|
Spaca3
|
sperm acrosome associated 3 |
| chr1_+_99648658 | 1.59 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr6_-_124429701 | 1.58 |
ENSRNOT00000037902
ENSRNOT00000080950 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr8_-_21384131 | 1.56 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chrX_-_45000363 | 1.55 |
ENSRNOT00000035538
|
Cldn34e
|
claudin 34E |
| chr6_-_26051229 | 1.44 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr3_+_18315320 | 1.44 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr16_+_7088476 | 1.41 |
ENSRNOT00000024624
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr10_+_10774639 | 1.26 |
ENSRNOT00000061208
|
Sept12
|
septin 12 |
| chr20_+_3827086 | 1.24 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr3_+_176865156 | 1.19 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr6_-_92527711 | 1.17 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr1_-_100746021 | 0.97 |
ENSRNOT00000037973
|
Zfp473
|
zinc finger protein 473 |
| chr1_+_255040426 | 0.94 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr2_+_178117466 | 0.92 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr11_-_84037938 | 0.81 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr10_+_14216155 | 0.70 |
ENSRNOT00000020192
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr4_+_147514120 | 0.61 |
ENSRNOT00000013146
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chr2_+_203768117 | 0.48 |
ENSRNOT00000080176
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_+_88182155 | 0.43 |
ENSRNOT00000083176
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr2_-_62887048 | 0.17 |
ENSRNOT00000039234
|
RGD1306502
|
similar to hypothetical protein FLJ11193 |
| chr15_+_34296922 | 0.15 |
ENSRNOT00000026430
|
Rec8
|
REC8 meiotic recombination protein |
| chr3_-_72219246 | 0.13 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr12_-_21746236 | 0.12 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rara
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 26.9 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 3.2 | 25.8 | GO:0090240 | negative regulation of cell adhesion mediated by integrin(GO:0033629) positive regulation of histone H4 acetylation(GO:0090240) |
| 2.3 | 18.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 2.0 | 2.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.0 | 7.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.7 | 13.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.6 | 9.9 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 1.6 | 6.4 | GO:0050893 | sensory processing(GO:0050893) |
| 1.3 | 14.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 1.3 | 5.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 1.2 | 6.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 1.0 | 8.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 1.0 | 8.0 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.8 | 3.4 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.8 | 14.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.7 | 11.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.7 | 7.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.6 | 20.9 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.5 | 7.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 2.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.3 | 3.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 19.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.3 | 2.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 11.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 11.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 1.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 3.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 3.9 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 9.0 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 2.6 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 4.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 2.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 4.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.9 | 18.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 2.0 | GO:0044307 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.4 | 6.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 9.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 10.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 3.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 26.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 12.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 12.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 3.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 11.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0097227 | septin complex(GO:0031105) sperm annulus(GO:0097227) |
| 0.1 | 8.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 5.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 6.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 8.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 29.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 21.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 16.5 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.0 | 14.0 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.6 | 11.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.6 | 20.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.9 | 7.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.6 | 2.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 11.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.5 | 2.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.5 | 2.7 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 8.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 3.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 11.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 11.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 25.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.3 | 11.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 3.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 7.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 7.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 4.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 10.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 6.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 4.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 4.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 8.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 8.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 7.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 54.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 17.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 7.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 11.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 13.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 20.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.7 | 18.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 7.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 10.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 4.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |