Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Ptf1a
Z-value: 0.89
Transcription factors associated with Ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ptf1a
|
ENSRNOG00000016902 | pancreas specific transcription factor, 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ptf1a | rn6_v1_chr17_+_86199623_86199623 | 0.49 | 6.8e-21 | Click! |
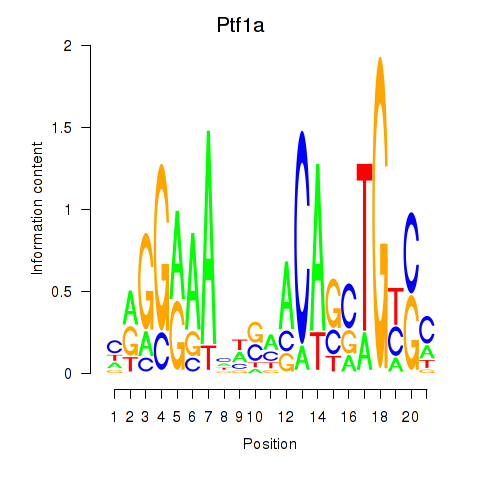
Activity profile of Ptf1a motif
Sorted Z-values of Ptf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_43911057 | 59.92 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr4_+_70755795 | 46.48 |
ENSRNOT00000043527
|
LOC683849
|
similar to Anionic trypsin II precursor (Pretrypsinogen II) |
| chr4_+_70689737 | 44.54 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr4_+_70614524 | 36.92 |
ENSRNOT00000041100
|
Prss3
|
protease, serine 3 |
| chr7_+_70807867 | 33.90 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr1_-_255376833 | 31.06 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr10_-_95934345 | 30.16 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr4_-_70747226 | 29.38 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr2_-_173668555 | 29.26 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr4_-_70628470 | 26.15 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr8_-_111777602 | 18.50 |
ENSRNOT00000052317
ENSRNOT00000083855 |
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr4_-_30338679 | 17.25 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr18_+_25163561 | 14.91 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr10_+_70262361 | 14.78 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr7_-_71226150 | 14.59 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr18_-_25314047 | 13.49 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr18_+_24708115 | 13.07 |
ENSRNOT00000061054
|
Lims2
|
LIM zinc finger domain containing 2 |
| chr4_+_115712850 | 12.87 |
ENSRNOT00000085765
|
Dysf
|
dysferlin |
| chr4_+_115713004 | 12.79 |
ENSRNOT00000064536
|
Dysf
|
dysferlin |
| chr4_+_100166863 | 12.64 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr20_-_5754773 | 12.45 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr11_-_64952687 | 12.00 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr16_-_75637789 | 11.29 |
ENSRNOT00000058029
|
Defb4
|
defensin beta 4 |
| chr3_+_58632476 | 10.57 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr10_+_47412582 | 9.95 |
ENSRNOT00000003139
|
Tnfrsf13b
|
TNF receptor superfamily member 13B |
| chr9_+_100156154 | 9.31 |
ENSRNOT00000005935
|
Aqp12a
|
aquaporin 12A |
| chr10_-_97896525 | 8.64 |
ENSRNOT00000005172
|
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr1_+_201055644 | 8.56 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_-_113228439 | 8.49 |
ENSRNOT00000034672
|
Col22a1
|
collagen type XXII alpha 1 chain |
| chr19_+_487723 | 7.91 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr6_-_141291347 | 7.80 |
ENSRNOT00000008333
|
AABR07065789.1
|
|
| chr14_+_17228856 | 7.61 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr4_+_102876038 | 7.53 |
ENSRNOT00000085205
|
AABR07061022.3
|
|
| chr6_+_127946686 | 7.47 |
ENSRNOT00000082680
|
LOC500712
|
Ab1-233 |
| chr14_-_86706626 | 7.34 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr4_+_93888502 | 7.05 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr8_+_116332796 | 6.38 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr4_-_156276304 | 6.27 |
ENSRNOT00000078725
|
Clec4e
|
C-type lectin domain family 4, member E |
| chrX_+_16946207 | 6.22 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr17_-_77304530 | 6.19 |
ENSRNOT00000024362
|
Phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr16_-_88347369 | 6.03 |
ENSRNOT00000019797
|
Pdha1l1
|
pyruvate dehydrogenase (lipoamide) alpha 1-like 1 |
| chr4_-_119927705 | 5.50 |
ENSRNOT00000077624
|
Rab7a
|
RAB7A, member RAS oncogene family |
| chr16_-_8350067 | 5.33 |
ENSRNOT00000026932
|
LOC100362432
|
mitochondrial import inner membrane translocase subunit Tim23-like |
| chr6_-_140572023 | 5.14 |
ENSRNOT00000072338
|
AABR07065772.1
|
|
| chr8_-_22874637 | 5.02 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr5_-_164747083 | 4.96 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr7_-_139483997 | 4.89 |
ENSRNOT00000086062
|
Col2a1
|
collagen type II alpha 1 chain |
| chr20_-_7943575 | 4.71 |
ENSRNOT00000066897
|
Tulp1
|
tubby like protein 1 |
| chr9_+_94928489 | 4.70 |
ENSRNOT00000087366
ENSRNOT00000085770 ENSRNOT00000024733 |
Sag
|
S-antigen visual arrestin |
| chr4_-_148690998 | 4.66 |
ENSRNOT00000052024
|
Olr831
|
olfactory receptor 831 |
| chr1_-_282251257 | 4.61 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr2_+_95320283 | 4.60 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr11_+_28724341 | 4.46 |
ENSRNOT00000049858
|
LOC100363287
|
RIKEN cDNA 2310034C09-like |
| chr17_-_42927704 | 4.39 |
ENSRNOT00000087291
|
Prl3d4
|
prolactin family 3, subfamily d, member 4 |
| chr5_+_113725717 | 4.34 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr10_+_89251370 | 4.26 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr8_+_20265891 | 4.24 |
ENSRNOT00000048326
|
Olr1166
|
olfactory receptor 1166 |
| chr8_-_126495347 | 4.24 |
ENSRNOT00000013467
|
Cmc1
|
C-x(9)-C motif containing 1 |
| chr7_-_143552588 | 4.23 |
ENSRNOT00000086317
ENSRNOT00000092619 ENSRNOT00000092138 |
Krt78
|
keratin 78 |
| chr17_-_39278695 | 4.14 |
ENSRNOT00000082821
|
LOC103689989
|
prolactin-3D4 |
| chr4_-_162025090 | 4.00 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chr3_+_19174027 | 3.99 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr6_+_128152343 | 3.97 |
ENSRNOT00000085867
|
Gm1000
|
predicted gene 10000 |
| chr3_+_67849966 | 3.79 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr1_+_226030938 | 3.77 |
ENSRNOT00000030919
|
Fth1
|
ferritin heavy chain 1 |
| chr16_-_64716224 | 3.76 |
ENSRNOT00000071166
|
Fut10
|
fucosyltransferase 10 |
| chr4_-_103636632 | 3.70 |
ENSRNOT00000086523
|
AABR07061077.1
|
|
| chr16_-_9430743 | 3.53 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr1_+_170887363 | 3.23 |
ENSRNOT00000036261
|
Olr215
|
olfactory receptor 215 |
| chr19_+_25946979 | 3.23 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr20_+_5008508 | 3.21 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr12_+_19680712 | 3.10 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr10_+_103349172 | 3.01 |
ENSRNOT00000035963
|
Gpr142
|
G protein-coupled receptor 142 |
| chr3_+_43293185 | 2.93 |
ENSRNOT00000090323
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr4_+_148664950 | 2.91 |
ENSRNOT00000017843
|
Olr830
|
olfactory receptor 830 |
| chr8_+_43733545 | 2.83 |
ENSRNOT00000082000
|
Olr1326
|
olfactory receptor 1326 |
| chr12_-_41625403 | 2.70 |
ENSRNOT00000001876
|
Sds
|
serine dehydratase |
| chr1_-_213981317 | 2.69 |
ENSRNOT00000086238
ENSRNOT00000054869 |
Slc25a22
|
solute carrier family 25 member 22 |
| chr9_+_43259709 | 2.68 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr15_+_28319136 | 2.57 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr5_+_69784806 | 2.57 |
ENSRNOT00000024722
|
Olr853
|
olfactory receptor 853 |
| chr4_-_115516296 | 2.54 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr7_-_60294796 | 2.47 |
ENSRNOT00000007580
|
Yeats4
|
YEATS domain containing 4 |
| chr3_+_103192683 | 2.46 |
ENSRNOT00000037551
|
LOC100363452
|
ribosomal protein S8-like |
| chr1_-_98046770 | 2.38 |
ENSRNOT00000056528
|
Grb2
|
growth factor receptor bound protein 2 |
| chr5_-_128333805 | 2.38 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr1_-_215330492 | 2.33 |
ENSRNOT00000068147
ENSRNOT00000042047 |
AABR07006049.1
|
|
| chr1_+_80973739 | 2.29 |
ENSRNOT00000026088
ENSRNOT00000081983 |
AC118165.1
|
|
| chr10_+_44958544 | 2.12 |
ENSRNOT00000072282
|
Olr1439
|
olfactory receptor 1439 |
| chr11_-_28736000 | 2.12 |
ENSRNOT00000064058
|
Krtap14l
|
keratin associated protein 14 like |
| chr1_+_229722768 | 2.11 |
ENSRNOT00000017126
|
Olr339
|
olfactory receptor 339 |
| chr1_-_82580007 | 2.10 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr14_-_18634309 | 2.06 |
ENSRNOT00000003720
|
Epgn
|
epithelial mitogen |
| chr15_-_27077160 | 1.99 |
ENSRNOT00000016721
|
Olr1611
|
olfactory receptor 1611 |
| chr5_-_148444188 | 1.87 |
ENSRNOT00000033929
|
RGD1562036
|
similar to ornithine decarboxylase |
| chr16_-_52127591 | 1.86 |
ENSRNOT00000033652
|
Triml2
|
tripartite motif family-like 2 |
| chr3_-_77170085 | 1.77 |
ENSRNOT00000042667
|
Olr654
|
olfactory receptor 654 |
| chr1_-_215350161 | 1.69 |
ENSRNOT00000081144
ENSRNOT00000064050 |
AABR07006049.1
|
|
| chr3_-_101547478 | 1.63 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr8_+_64531594 | 1.49 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr8_+_22035256 | 1.41 |
ENSRNOT00000028066
|
Icam1
|
intercellular adhesion molecule 1 |
| chr19_-_54761670 | 1.39 |
ENSRNOT00000025848
|
Ca5a
|
carbonic anhydrase 5A |
| chr18_+_15738809 | 1.38 |
ENSRNOT00000075444
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_-_12571316 | 1.37 |
ENSRNOT00000074440
|
Olr1372
|
olfactory receptor 1372 |
| chr1_-_99632845 | 1.36 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr4_-_148437961 | 1.30 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr1_-_98501249 | 1.18 |
ENSRNOT00000023858
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr7_+_130357643 | 1.14 |
ENSRNOT00000040769
|
Klhdc7b
|
kelch domain containing 7B |
| chr7_-_130405347 | 1.09 |
ENSRNOT00000013985
|
Cpt1b
|
carnitine palmitoyltransferase 1B |
| chr20_-_848229 | 1.08 |
ENSRNOT00000000974
|
Olr1694
|
olfactory receptor 1694 |
| chr1_+_77541359 | 1.07 |
ENSRNOT00000049028
|
LOC103690015
|
40S ribosomal protein S19-like |
| chr1_+_170796271 | 1.06 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr16_-_38063821 | 1.06 |
ENSRNOT00000077452
ENSRNOT00000089533 |
Glra3
|
glycine receptor, alpha 3 |
| chr1_+_79396578 | 1.03 |
ENSRNOT00000023686
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr3_+_153402548 | 0.97 |
ENSRNOT00000091273
|
Rpn2
|
ribophorin II |
| chr10_+_54126486 | 0.97 |
ENSRNOT00000078820
|
Gas7
|
growth arrest specific 7 |
| chr10_+_87788458 | 0.95 |
ENSRNOT00000042020
|
LOC100910814
|
keratin-associated protein 9-1-like |
| chr17_-_62299655 | 0.94 |
ENSRNOT00000024855
|
Gjd4
|
gap junction protein, delta 4 |
| chr6_+_38474804 | 0.93 |
ENSRNOT00000009779
|
Nbas
|
neuroblastoma amplified sequence |
| chr1_-_167971151 | 0.92 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr4_-_103403976 | 0.91 |
ENSRNOT00000088153
|
AABR07061057.3
|
|
| chr1_-_170927957 | 0.90 |
ENSRNOT00000049686
|
Olr218
|
olfactory receptor 218 |
| chr19_+_27464937 | 0.83 |
ENSRNOT00000024020
|
Vps35
|
VPS35 retromer complex component |
| chr20_+_1944801 | 0.80 |
ENSRNOT00000075060
|
Olr1750
|
olfactory receptor 1750 |
| chr5_+_137757938 | 0.80 |
ENSRNOT00000073594
|
LOC100912491
|
olfactory receptor 2A2-like |
| chr20_-_11644166 | 0.79 |
ENSRNOT00000073009
|
LOC100361739
|
keratin associated protein 12-1-like |
| chr5_-_6083083 | 0.77 |
ENSRNOT00000072411
|
Sulf1
|
sulfatase 1 |
| chr8_+_56451240 | 0.73 |
ENSRNOT00000085278
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr7_+_136037230 | 0.72 |
ENSRNOT00000071423
|
AABR07058745.1
|
|
| chr3_+_20893187 | 0.71 |
ENSRNOT00000070947
|
Olr415
|
olfactory receptor 415 |
| chr19_-_10581842 | 0.68 |
ENSRNOT00000021639
|
Polr2c
|
RNA polymerase II subunit C |
| chr11_+_74834050 | 0.57 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr5_-_137515690 | 0.56 |
ENSRNOT00000049593
|
LOC100912421
|
olfactory receptor 2D2-like |
| chr1_-_73961432 | 0.55 |
ENSRNOT00000035435
|
Nlrp4b
|
NLR family, pyrin domain containing 4B |
| chr1_-_54748763 | 0.50 |
ENSRNOT00000074549
|
LOC100911027
|
protein MAL2-like |
| chr7_+_141702038 | 0.47 |
ENSRNOT00000086577
|
Dip2b
|
disco-interacting protein 2 homolog B |
| chr3_+_160852164 | 0.43 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr13_+_96111800 | 0.41 |
ENSRNOT00000005967
|
Desi2
|
desumoylating isopeptidase 2 |
| chr20_+_3367208 | 0.39 |
ENSRNOT00000091149
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr1_-_170763377 | 0.35 |
ENSRNOT00000041335
|
Olr209
|
olfactory receptor 209 |
| chr2_+_112868707 | 0.30 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr15_-_41316203 | 0.30 |
ENSRNOT00000065261
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr9_+_16302578 | 0.28 |
ENSRNOT00000021762
|
Gltscr1l
|
GLTSCR1-like |
| chr1_-_167698263 | 0.26 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr3_-_78409263 | 0.23 |
ENSRNOT00000041001
|
Olr704
|
olfactory receptor 704 |
| chr1_+_259739955 | 0.21 |
ENSRNOT00000054719
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr16_-_37908173 | 0.16 |
ENSRNOT00000075026
|
Glra3
|
glycine receptor, alpha 3 |
| chr10_-_45579029 | 0.13 |
ENSRNOT00000080028
|
Arf1
|
ADP-ribosylation factor 1 |
| chr10_+_87819150 | 0.13 |
ENSRNOT00000073764
ENSRNOT00000064626 |
LOC680442
|
hypothetical protein LOC680442 |
| chr2_+_87957363 | 0.06 |
ENSRNOT00000047881
|
LOC499565
|
LRRGT00038 |
| chr10_-_87753668 | 0.06 |
ENSRNOT00000066758
ENSRNOT00000047104 |
Krtap4-3
|
keratin associated protein 4-3 |
| chr10_-_12674077 | 0.02 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr10_-_60275762 | 0.02 |
ENSRNOT00000074380
|
Olr1498
|
olfactory receptor 1498 |
| chr7_-_11724843 | 0.01 |
ENSRNOT00000084144
|
Sppl2b
|
signal peptide peptidase-like 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ptf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 4.0 | 12.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 3.5 | 10.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.9 | 17.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.6 | 31.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 2.5 | 12.6 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.1 | 6.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 2.0 | 25.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.8 | 5.5 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 1.7 | 5.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.6 | 13.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.6 | 6.4 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.5 | 4.6 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.4 | 4.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.2 | 187.9 | GO:0007586 | digestion(GO:0007586) |
| 1.0 | 4.9 | GO:0071306 | cellular response to vitamin E(GO:0071306) |
| 0.9 | 4.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.8 | 33.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.8 | 30.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.8 | 4.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.7 | 2.7 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.7 | 2.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.7 | 8.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.6 | 2.9 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.6 | 8.6 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.5 | 2.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.5 | 7.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 10.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.4 | 6.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.4 | 12.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.4 | 3.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.4 | 4.4 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.4 | 1.4 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.3 | 1.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.3 | 1.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 3.8 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of protein catabolic process in the vacuole(GO:1904350) lysosomal protein catabolic process(GO:1905146) |
| 0.3 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 3.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 4.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 11.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 14.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 0.9 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.2 | 0.9 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 2.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 3.8 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 2.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 2.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 2.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 7.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.2 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.1 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 3.9 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.1 | 2.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 8.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 22.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 5.9 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 31.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 2.1 | 6.4 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 2.1 | 10.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.5 | 14.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.3 | 18.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.8 | 8.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.7 | 30.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 2.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.6 | 2.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 5.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.4 | 4.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.2 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.4 | 4.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 25.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 3.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 0.9 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 0.8 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 9.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 5.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.1 | GO:0033643 | host cell part(GO:0033643) |
| 0.2 | 4.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 236.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 12.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 2.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 7.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 9.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 6.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.6 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 2.8 | 11.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.7 | 5.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 1.5 | 4.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.5 | 7.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.4 | 12.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.3 | 6.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.3 | 3.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.2 | 4.7 | GO:0002046 | opsin binding(GO:0002046) |
| 1.2 | 4.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.0 | 2.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.9 | 2.7 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.8 | 4.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.8 | 180.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.7 | 4.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.7 | 14.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 3.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 22.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 30.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.5 | 2.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 4.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 3.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.4 | 1.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.4 | 31.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 1.2 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.4 | 4.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 14.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 1.9 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.4 | 25.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 8.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 6.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.3 | 1.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 2.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 29.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 2.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 5.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 18.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 8.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 4.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 8.1 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 8.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.1 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 22.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 100.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 14.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 13.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 10.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 13.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 2.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 7.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 4.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 5.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 18.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 44.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.4 | 38.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 10.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 6.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 6.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 18.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.3 | 6.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 8.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 7.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 5.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.9 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |