Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
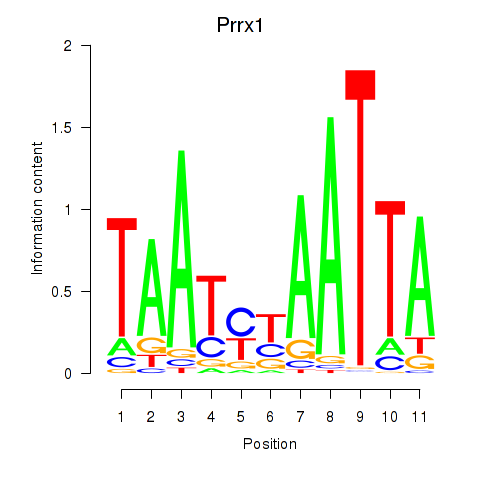
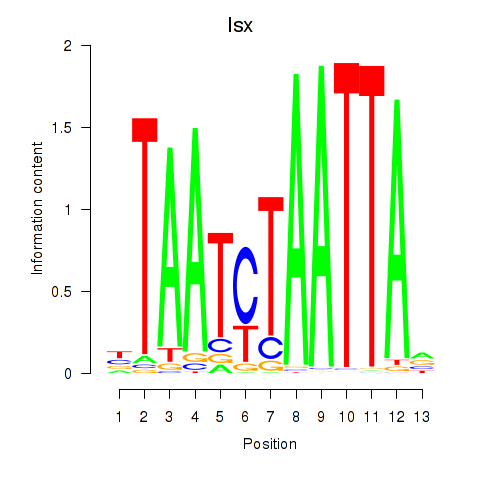
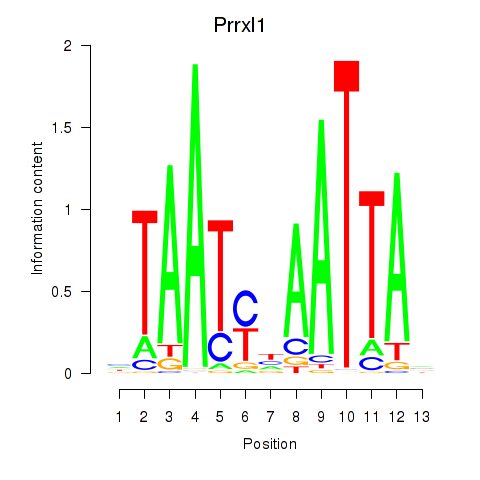
Results for Prrx1_Isx_Prrxl1
Z-value: 0.81
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prrx1
|
ENSRNOG00000003720 | paired related homeobox 1 |
|
Isx
|
ENSRNOG00000039808 | intestine-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Drgx | rn6_v1_chr16_+_8823872_8823872 | 0.33 | 1.9e-09 | Click! |
| Prrx1 | rn6_v1_chr13_-_81214494_81214546 | 0.31 | 2.4e-08 | Click! |
| Isx | rn6_v1_chr19_+_14198899_14198899 | 0.03 | 6.3e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_198303168 | 21.79 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr5_+_118574801 | 20.31 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr18_-_24057917 | 20.30 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr4_-_55011415 | 19.23 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr4_-_61720956 | 18.38 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr20_+_31339787 | 17.96 |
ENSRNOT00000082463
|
Aifm2
|
apoptosis inducing factor, mitochondria associated 2 |
| chr7_+_59200918 | 17.63 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_6305033 | 16.74 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr4_+_70252366 | 16.33 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr17_+_63635086 | 16.25 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr8_+_82038967 | 16.23 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr17_+_81922329 | 15.38 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr14_+_69800156 | 14.93 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr6_-_14523961 | 14.87 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr6_-_2311781 | 14.55 |
ENSRNOT00000084171
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr7_-_52404774 | 13.69 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr7_-_29171783 | 13.54 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr11_+_20474483 | 13.38 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr7_-_107768072 | 13.24 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr18_+_30398113 | 13.05 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr2_+_266315036 | 12.75 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr10_+_56381813 | 12.71 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr3_-_21904133 | 12.70 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr2_-_156807305 | 12.61 |
ENSRNOT00000080578
|
AABR07011031.1
|
uncharacterized protein LOC302022 |
| chr8_+_126975833 | 12.31 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr8_-_102149912 | 12.30 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr2_-_40386669 | 12.03 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr3_-_134696654 | 11.98 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr9_+_73378057 | 11.75 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_5709236 | 11.72 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr10_-_15928169 | 11.56 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr5_+_103251986 | 11.55 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr3_+_173799833 | 11.36 |
ENSRNOT00000081235
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr6_+_8284878 | 11.26 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr10_+_77537340 | 11.05 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr12_-_46493203 | 10.89 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chrX_+_14019961 | 10.85 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr2_-_88763733 | 10.85 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chrX_+_84064427 | 10.82 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_-_26056818 | 10.78 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr14_-_66978499 | 10.73 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr3_+_56056925 | 10.34 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr20_+_19325121 | 10.29 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr4_+_2055615 | 10.25 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr1_+_234363994 | 10.22 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr6_-_127319362 | 10.10 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr2_-_117454769 | 10.10 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr2_-_158156444 | 10.01 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_9956370 | 10.00 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr13_-_76049363 | 9.88 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr13_+_78979321 | 9.73 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr17_-_84247038 | 9.51 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr5_-_57896475 | 9.44 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chrX_-_40086870 | 9.39 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr1_+_60884761 | 9.36 |
ENSRNOT00000088689
|
LOC100365363
|
vomeronasal 1 receptor, F4-like |
| chr4_-_17440506 | 9.12 |
ENSRNOT00000080931
|
Sema3e
|
semaphorin 3E |
| chr8_+_117062884 | 9.03 |
ENSRNOT00000082452
ENSRNOT00000071540 |
Nicn1
|
nicolin 1 |
| chrM_+_9870 | 9.01 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_-_64364101 | 8.93 |
ENSRNOT00000089065
|
Zfp385b
|
zinc finger protein 385B |
| chr2_-_149444548 | 8.68 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr15_+_62406873 | 8.62 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr15_+_43298794 | 8.44 |
ENSRNOT00000012736
|
Adra1a
|
adrenoceptor alpha 1A |
| chr6_-_114476723 | 8.43 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr2_-_231648122 | 8.32 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr10_+_46722109 | 8.28 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chr7_-_75288365 | 8.19 |
ENSRNOT00000036904
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr2_-_38110567 | 8.14 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chrM_+_10160 | 8.13 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_-_53802658 | 7.84 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr10_-_66848388 | 7.80 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr19_-_24614019 | 7.77 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr1_-_67134827 | 7.59 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr8_-_132753145 | 7.54 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chr9_+_84203072 | 7.38 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chrX_-_77559348 | 7.36 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr14_-_3300200 | 7.21 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr4_+_88328061 | 7.09 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr1_+_53874860 | 7.04 |
ENSRNOT00000090486
|
Tcte2
|
t-complex-associated testis expressed 2 |
| chr8_+_64489942 | 7.04 |
ENSRNOT00000083666
|
Pkm
|
pyruvate kinase, muscle |
| chr10_+_97771264 | 6.99 |
ENSRNOT00000005257
|
Arsg
|
arylsulfatase G |
| chr10_-_16731898 | 6.94 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr12_-_54885 | 6.70 |
ENSRNOT00000090447
|
AABR07034833.1
|
|
| chrX_-_142164220 | 6.68 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr17_-_14627937 | 6.59 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr16_-_39476384 | 6.57 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr4_+_88184956 | 6.54 |
ENSRNOT00000077129
|
Vom1r83
|
vomeronasal 1 receptor 83 |
| chr18_+_30474947 | 6.48 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr4_+_88271061 | 6.47 |
ENSRNOT00000087374
|
Vom1r86
|
vomeronasal 1 receptor 86 |
| chr4_-_15859132 | 6.43 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr13_+_94888078 | 6.31 |
ENSRNOT00000077995
ENSRNOT00000005647 ENSRNOT00000077345 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr18_-_32207749 | 6.26 |
ENSRNOT00000090882
ENSRNOT00000018843 |
Arhgap26
|
Rho GTPase activating protein 26 |
| chr18_+_31444472 | 6.21 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr6_+_29977797 | 6.13 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr10_-_91661558 | 6.13 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr3_+_155160481 | 6.03 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr8_-_47094352 | 6.02 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr18_+_30487264 | 6.00 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr1_+_188420461 | 5.96 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr17_+_16340226 | 5.93 |
ENSRNOT00000089140
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_63803915 | 5.86 |
ENSRNOT00000076938
|
Maged1
|
MAGE family member D1 |
| chr20_+_20378861 | 5.83 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chr2_+_242634399 | 5.77 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr7_-_76294663 | 5.77 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr7_-_143353925 | 5.74 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chrX_+_118197217 | 5.70 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr3_+_8547349 | 5.58 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr9_+_60039297 | 5.58 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr9_+_111597037 | 5.50 |
ENSRNOT00000021758
|
Fer
|
FER tyrosine kinase |
| chrM_+_9451 | 5.47 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_-_138683318 | 5.39 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr14_+_39663421 | 5.20 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr1_+_66959610 | 5.19 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr3_+_81134505 | 5.18 |
ENSRNOT00000067664
|
Phf21a
|
PHD finger protein 21A |
| chr16_+_78539489 | 5.17 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr18_+_63203063 | 5.16 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr10_+_92289107 | 5.12 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr8_+_26432514 | 5.12 |
ENSRNOT00000081574
|
Sept7
|
septin 7 |
| chr15_+_56666012 | 5.12 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr1_+_61786900 | 4.98 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr8_-_83280888 | 4.97 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chrX_+_92131209 | 4.90 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr5_-_147412705 | 4.89 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr2_-_57935334 | 4.89 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr7_-_70476340 | 4.86 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chrX_-_121731543 | 4.80 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr5_+_6373583 | 4.79 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr9_+_73418607 | 4.77 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr2_+_22000106 | 4.75 |
ENSRNOT00000061812
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr10_-_86645529 | 4.74 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr3_-_25212049 | 4.74 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr7_+_380741 | 4.74 |
ENSRNOT00000091013
|
LOC102554748
|
nidogen-2-like |
| chr13_-_80775230 | 4.73 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr3_-_124724252 | 4.72 |
ENSRNOT00000028885
|
Slc23a2
|
solute carrier family 23 member 2 |
| chr2_-_257546799 | 4.69 |
ENSRNOT00000089370
ENSRNOT00000090367 |
Miga1
|
mitoguardin 1 |
| chr12_-_2174131 | 4.67 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr9_+_62291809 | 4.55 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr4_+_155321553 | 4.51 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr12_+_39423596 | 4.49 |
ENSRNOT00000066952
|
Ift81
|
intraflagellar transport 81 |
| chr7_+_23403891 | 4.46 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chrX_+_144994139 | 4.45 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr11_-_72814850 | 4.44 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chrX_-_83151511 | 4.44 |
ENSRNOT00000057378
|
Hdx
|
highly divergent homeobox |
| chr7_+_27174882 | 4.43 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr10_-_36323055 | 4.42 |
ENSRNOT00000081026
|
Zfp354c
|
zinc finger protein 354C |
| chr4_+_88119838 | 4.39 |
ENSRNOT00000073173
|
Vom1r82
|
vomeronasal 1 receptor 82 |
| chrM_-_14061 | 4.38 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr11_+_20471764 | 4.38 |
ENSRNOT00000078127
|
Ncam2
|
neural cell adhesion molecule 2 |
| chr6_+_48708368 | 4.36 |
ENSRNOT00000005905
|
Myt1l
|
myelin transcription factor 1-like |
| chr8_-_84835060 | 4.34 |
ENSRNOT00000007867
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr1_-_67065797 | 4.28 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr17_-_61332391 | 4.23 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr17_-_87826421 | 4.23 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr1_-_84950210 | 4.20 |
ENSRNOT00000083050
|
AABR07002784.1
|
|
| chr3_+_56862691 | 4.19 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr2_+_217963456 | 4.19 |
ENSRNOT00000024243
|
Olfm3
|
olfactomedin 3 |
| chr8_+_64531594 | 4.14 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr7_+_117456039 | 4.06 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr18_+_30496318 | 4.03 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr5_-_115167986 | 4.02 |
ENSRNOT00000079786
|
LOC100912642
|
cytochrome P450 2J3-like |
| chrX_-_123092217 | 4.01 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr3_-_76696107 | 4.00 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr3_-_15278645 | 3.99 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chrM_+_14136 | 3.98 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr3_+_48082935 | 3.92 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr9_-_61810417 | 3.90 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr8_-_18762922 | 3.89 |
ENSRNOT00000008423
|
Olr1122
|
olfactory receptor 1122 |
| chr2_+_143656793 | 3.87 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr2_+_58448917 | 3.87 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr10_+_17327275 | 3.86 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr7_-_29152442 | 3.85 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr5_+_154868534 | 3.84 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr18_+_32336102 | 3.77 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr8_-_39551700 | 3.77 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr6_+_64649194 | 3.75 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr2_+_200793571 | 3.74 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chrX_-_37003642 | 3.72 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr15_+_4209703 | 3.72 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr13_-_86671515 | 3.72 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr9_+_10941613 | 3.71 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr20_+_37876650 | 3.67 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr6_+_48452369 | 3.66 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr3_+_76468294 | 3.63 |
ENSRNOT00000037779
|
Olr619
|
olfactory receptor 619 |
| chr16_+_32457521 | 3.63 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr14_-_28967980 | 3.60 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr2_-_158156150 | 3.58 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_-_81693000 | 3.57 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chrX_+_908044 | 3.56 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr20_-_10013190 | 3.54 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr12_-_52658275 | 3.54 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr5_-_167696348 | 3.50 |
ENSRNOT00000024581
|
Slc45a1
|
solute carrier family 45, member 1 |
| chr18_+_30036887 | 3.48 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr16_-_74496731 | 3.46 |
ENSRNOT00000015790
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr14_+_42015347 | 3.45 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr1_-_22281788 | 3.45 |
ENSRNOT00000021318
|
Stx7
|
syntaxin 7 |
| chrM_+_7006 | 3.42 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_+_80670140 | 3.42 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr5_-_25721072 | 3.41 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr17_-_15566332 | 3.39 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr8_-_8524643 | 3.39 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr3_+_47677720 | 3.38 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 5.1 | 15.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 4.9 | 14.6 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 4.4 | 17.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 4.2 | 12.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 4.1 | 16.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 3.9 | 11.7 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 3.8 | 19.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 3.6 | 10.8 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 3.6 | 10.7 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 3.2 | 9.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.9 | 20.3 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 2.9 | 8.7 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 2.8 | 8.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 2.4 | 12.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.3 | 6.9 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 2.1 | 8.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.0 | 6.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 2.0 | 6.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.8 | 9.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.7 | 19.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.7 | 17.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.7 | 5.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 1.6 | 6.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.6 | 11.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.6 | 4.7 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.6 | 7.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.5 | 5.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.4 | 8.4 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 1.4 | 5.5 | GO:0038109 | Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 1.3 | 12.0 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 1.3 | 3.9 | GO:1904209 | cellular response to vitamin K(GO:0071307) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.3 | 3.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.2 | 3.7 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 1.2 | 3.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.2 | 3.7 | GO:0010643 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 1.2 | 13.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 1.2 | 3.6 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.2 | 4.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 1.2 | 8.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.2 | 11.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 1.2 | 3.5 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.1 | 10.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 1.1 | 3.3 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.1 | 1.1 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) |
| 1.1 | 3.2 | GO:1903544 | response to butyrate(GO:1903544) |
| 1.0 | 12.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.0 | 6.7 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.9 | 3.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 2.8 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.9 | 2.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.9 | 3.6 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.9 | 6.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.9 | 2.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.9 | 3.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 8.6 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.8 | 4.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.8 | 2.5 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.8 | 2.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.8 | 2.4 | GO:0015692 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 0.8 | 7.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.8 | 4.6 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.8 | 4.5 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 | 6.4 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.7 | 7.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.7 | 5.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 4.0 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.7 | 3.3 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.7 | 23.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.6 | 4.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.6 | 5.1 | GO:1902474 | negative regulation of mitochondrial fission(GO:0090258) regulation of cellular response to heat(GO:1900034) regulation of protein localization to synapse(GO:1902473) positive regulation of protein localization to synapse(GO:1902474) |
| 0.6 | 12.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.6 | 3.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 7.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.6 | 5.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.6 | 4.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.6 | 2.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.6 | 1.7 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 1.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.6 | 1.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.6 | 3.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.6 | 2.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.6 | 2.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 17.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.5 | 2.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.5 | 5.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 4.2 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.5 | 1.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.5 | 2.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.5 | 10.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.5 | 2.4 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.5 | 3.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 10.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.5 | 2.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 5.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.5 | 1.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.5 | 1.4 | GO:0035425 | autocrine signaling(GO:0035425) paracrine signaling(GO:0038001) |
| 0.4 | 16.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 2.7 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.4 | 20.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.4 | 2.6 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.4 | 2.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 3.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 2.1 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.4 | 0.8 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.4 | 1.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.4 | 2.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 4.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 3.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.4 | 3.4 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.4 | 1.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.4 | 1.8 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.3 | 2.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 5.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.3 | 1.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 6.9 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 1.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.3 | 3.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 0.3 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.3 | 1.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.3 | 16.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.3 | 12.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.3 | 7.8 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.3 | 2.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 3.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.3 | 0.3 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.3 | 39.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.3 | 10.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.3 | 9.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 12.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.3 | 4.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 1.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 4.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 0.8 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.2 | 2.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 6.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 2.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 2.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 6.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 10.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 6.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.2 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 4.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 3.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.2 | 1.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.2 | 1.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 2.5 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 0.8 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 20.8 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.2 | 1.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 10.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.6 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 2.5 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.2 | 2.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 6.0 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 4.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 5.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 2.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 1.0 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.2 | 8.0 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.2 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.2 | 1.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.2 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.9 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.6 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 5.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.5 | GO:0070127 | alanyl-tRNA aminoacylation(GO:0006419) tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 3.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.7 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 2.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 1.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 3.9 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 1.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.6 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 1.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.7 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 1.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 5.9 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.6 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 5.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 5.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 4.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 5.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.8 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 1.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 2.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 8.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 1.0 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 5.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 1.6 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 10.6 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 2.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 2.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 5.3 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 5.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 2.2 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.5 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.0 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.0 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 4.2 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.4 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.8 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 5.1 | 20.3 | GO:0033503 | HULC complex(GO:0033503) |
| 3.7 | 18.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.9 | 11.7 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 2.6 | 23.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.9 | 9.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.8 | 21.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.6 | 4.7 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 1.4 | 17.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.3 | 5.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.1 | 3.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 1.0 | 5.1 | GO:0005940 | septin ring(GO:0005940) |
| 1.0 | 6.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.9 | 3.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.9 | 3.4 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.9 | 9.4 | GO:0002177 | manchette(GO:0002177) |
| 0.8 | 2.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.8 | 2.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.8 | 33.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 12.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.7 | 12.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 5.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.6 | 3.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 5.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 11.0 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 1.7 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.5 | 16.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.5 | 25.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.5 | 14.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 2.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 2.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 4.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 2.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 13.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 5.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 1.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 0.9 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 19.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 0.8 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.3 | 20.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 13.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 3.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 2.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 7.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 2.9 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 12.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 18.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 4.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 2.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 2.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 15.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 11.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 6.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 3.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 3.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 19.2 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 6.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 15.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 5.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 3.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 6.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 13.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 9.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 2.0 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 3.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 8.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 7.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 4.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 10.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 11.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 6.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 18.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 23.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 3.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 32.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 9.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 6.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 16.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 11.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 6.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 11.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 12.5 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 3.9 | 11.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 3.6 | 10.8 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 2.8 | 8.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.7 | 10.7 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 2.6 | 15.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 2.5 | 33.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 2.5 | 7.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.1 | 6.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 2.1 | 14.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.0 | 10.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.8 | 12.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.8 | 7.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.7 | 5.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.6 | 4.7 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.5 | 18.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.3 | 17.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.3 | 5.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.3 | 5.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.2 | 3.7 | GO:0052854 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 1.2 | 8.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.2 | 3.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.2 | 12.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 12.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.1 | 3.3 | GO:0099567 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 1.1 | 28.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.1 | 4.2 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 1.0 | 5.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.0 | 8.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 7.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.0 | 7.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.0 | 4.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.9 | 2.8 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.9 | 3.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.9 | 3.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 16.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.9 | 6.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.8 | 1.7 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.8 | 4.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.8 | 2.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.8 | 2.4 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.7 | 3.7 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.7 | 16.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.7 | 3.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 17.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.7 | 17.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.6 | 12.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 3.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.6 | 1.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.6 | 6.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 13.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.6 | 5.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 18.0 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.6 | 1.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.6 | 3.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.5 | 1.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 52.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.5 | 2.4 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.5 | 5.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 18.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 3.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 1.7 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.4 | 20.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 1.2 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.4 | 6.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 12.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 1.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 2.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 2.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 1.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 1.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.3 | 4.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 3.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 7.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 2.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 2.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 1.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 1.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 5.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 3.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 7.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 2.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 9.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.6 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 1.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.5 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 4.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 9.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.2 | 10.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 3.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 11.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 17.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 10.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 3.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 5.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 13.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 10.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 4.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 22.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 7.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 6.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.7 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 8.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 5.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 3.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 7.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 3.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 16.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 3.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 13.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 3.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 10.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 10.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 3.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 20.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.5 | 5.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 11.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 10.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 17.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 16.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 9.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 7.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 15.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 15.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 3.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 2.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 22.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 1.0 | 20.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.9 | 16.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.9 | 15.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.9 | 11.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.9 | 19.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 11.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.7 | 18.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.7 | 7.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.7 | 12.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.6 | 16.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 9.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.5 | 10.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 17.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 5.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 3.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 4.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 12.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 10.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 8.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 5.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 5.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 3.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 4.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 3.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.3 | 9.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 2.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 2.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 7.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 1.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 1.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 3.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 9.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 6.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 9.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 5.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 5.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 13.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 4.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |