Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
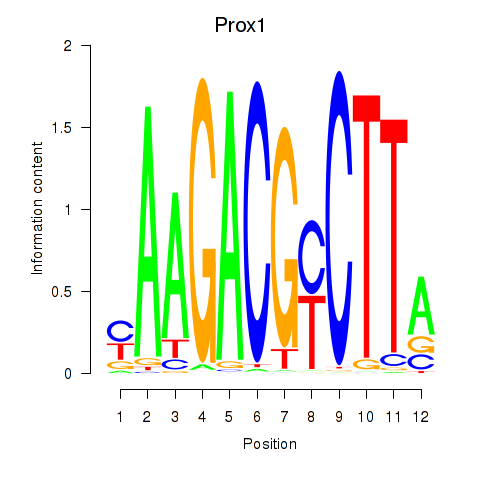
Results for Prox1
Z-value: 0.57
Transcription factors associated with Prox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prox1
|
ENSRNOG00000003694 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prox1 | rn6_v1_chr13_+_108382399_108382399 | -0.30 | 2.8e-08 | Click! |
Activity profile of Prox1 motif
Sorted Z-values of Prox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_42002819 | 18.04 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr13_+_51384389 | 12.39 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr13_+_51384562 | 12.27 |
ENSRNOT00000077928
|
Kdm5b
|
lysine demethylase 5B |
| chr5_+_151362019 | 11.95 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr1_-_84812486 | 10.86 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr1_-_80367313 | 10.37 |
ENSRNOT00000023392
ENSRNOT00000084882 |
Mark4
|
microtubule affinity regulating kinase 4 |
| chr5_+_147069616 | 9.95 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr8_-_5429581 | 8.88 |
ENSRNOT00000044174
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr4_-_166617143 | 8.15 |
ENSRNOT00000038077
|
AABR07062268.1
|
|
| chr5_+_25391115 | 8.10 |
ENSRNOT00000021612
|
Cdh17
|
cadherin 17 |
| chr19_+_9895121 | 7.47 |
ENSRNOT00000033953
|
Prss54
|
protease, serine, 54 |
| chr12_-_42492526 | 7.10 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr12_+_13810180 | 6.05 |
ENSRNOT00000084865
ENSRNOT00000038203 |
Tnrc18
|
trinucleotide repeat containing 18 |
| chr6_+_10533151 | 5.74 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr6_-_10592454 | 5.58 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr15_+_36865548 | 5.38 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr8_-_118182559 | 5.24 |
ENSRNOT00000084838
|
Dhx30
|
DExH-box helicase 30 |
| chr7_-_114848414 | 5.11 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr1_+_265904566 | 5.04 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr1_-_56982554 | 5.03 |
ENSRNOT00000047157
ENSRNOT00000078312 |
Tcte3
|
t-complex-associated testis expressed 3 |
| chr2_+_189413966 | 4.98 |
ENSRNOT00000056603
|
RGD1564171
|
RGD1564171 |
| chr6_+_51662224 | 4.95 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr14_+_91782354 | 4.61 |
ENSRNOT00000005902
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr19_-_49198073 | 4.43 |
ENSRNOT00000068440
|
Cdyl2
|
chromodomain Y-like 2 |
| chr7_+_144605058 | 4.19 |
ENSRNOT00000031404
|
Hoxc8
|
homeobox C8 |
| chr18_+_86307646 | 3.98 |
ENSRNOT00000091778
|
Cd226
|
CD226 molecule |
| chr12_-_19328637 | 3.85 |
ENSRNOT00000001829
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr7_-_126913585 | 3.80 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr6_+_6619076 | 3.74 |
ENSRNOT00000042307
ENSRNOT00000080013 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr5_+_149047681 | 3.73 |
ENSRNOT00000015198
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr10_-_57606171 | 3.70 |
ENSRNOT00000009015
|
Nup88
|
nucleoporin 88 |
| chr2_+_260039651 | 3.66 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr10_+_72147816 | 3.65 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chr17_-_76250537 | 3.61 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr1_-_198900375 | 3.49 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
| chr15_+_52451161 | 3.38 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr10_+_75620470 | 3.30 |
ENSRNOT00000013882
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr8_+_107229832 | 3.30 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr3_-_3295101 | 3.27 |
ENSRNOT00000051729
|
Sohlh1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr11_-_87769761 | 3.25 |
ENSRNOT00000052250
|
AABR07072266.1
|
|
| chr18_+_79773608 | 3.24 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr10_-_73787083 | 3.16 |
ENSRNOT00000035280
|
Med13
|
mediator complex subunit 13 |
| chr12_-_38053085 | 3.15 |
ENSRNOT00000034264
|
Hip1r
|
huntingtin interacting protein 1 related |
| chr4_+_25538497 | 3.04 |
ENSRNOT00000009264
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr5_+_139038134 | 2.96 |
ENSRNOT00000078155
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr16_+_19068783 | 2.92 |
ENSRNOT00000017424
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr8_+_106816152 | 2.91 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr3_-_23066658 | 2.88 |
ENSRNOT00000018751
|
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_-_26194198 | 2.71 |
ENSRNOT00000005917
|
Wdr83
|
WD repeat domain 83 |
| chr15_+_47470863 | 2.71 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr3_+_113131327 | 2.68 |
ENSRNOT00000018460
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr15_+_30550202 | 2.59 |
ENSRNOT00000087199
|
AABR07017745.3
|
|
| chr5_-_113532878 | 2.42 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr15_-_110046687 | 2.18 |
ENSRNOT00000057404
ENSRNOT00000006624 ENSRNOT00000089695 |
Nalcn
|
sodium leak channel, non-selective |
| chr1_-_255573377 | 2.16 |
ENSRNOT00000027780
|
Ubbp4
|
ubiquitin B pseudogene 4 |
| chr1_+_82473737 | 2.12 |
ENSRNOT00000028029
|
B9d2
|
B9 protein domain 2 |
| chr18_+_30880020 | 2.09 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr4_+_61924013 | 2.08 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr1_+_61374379 | 2.04 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr8_-_27973980 | 1.87 |
ENSRNOT00000037757
|
Bin2a
|
beta-galactosidase-like protein |
| chr6_+_72891725 | 1.81 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr15_-_28672530 | 1.55 |
ENSRNOT00000022593
ENSRNOT00000087722 |
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr7_+_97760134 | 1.47 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr20_-_10257044 | 1.45 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr20_+_1837976 | 1.39 |
ENSRNOT00000084564
|
Olr1742
|
olfactory receptor 1742 |
| chr10_+_11338306 | 1.25 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr9_+_117737611 | 1.22 |
ENSRNOT00000022396
|
Zfp161
|
zinc finger protein 161 |
| chr6_-_132608600 | 1.21 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr3_-_71295575 | 1.20 |
ENSRNOT00000040303
ENSRNOT00000083015 |
Zswim2
|
zinc finger, SWIM-type containing 2 |
| chr7_-_2819299 | 1.18 |
ENSRNOT00000045547
|
Slc39a5
|
solute carrier family 39 member 5 |
| chr10_+_10889488 | 1.15 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr4_+_21872856 | 1.10 |
ENSRNOT00000044810
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr2_+_86996497 | 1.06 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr10_-_102423084 | 1.00 |
ENSRNOT00000065145
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr4_-_117126822 | 0.95 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr2_-_185303610 | 0.73 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr8_+_116776494 | 0.67 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
| chr1_+_56982821 | 0.63 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr12_-_2007516 | 0.48 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr3_-_133122691 | 0.36 |
ENSRNOT00000083637
|
Tasp1
|
taspase 1 |
| chrX_+_156812064 | 0.29 |
ENSRNOT00000077142
|
Hcfc1
|
host cell factor C1 |
| chr2_-_140618405 | 0.24 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr13_+_71305548 | 0.21 |
ENSRNOT00000074864
|
LOC684709
|
similar to putative membrane protein Re9 |
| chr15_+_7871497 | 0.16 |
ENSRNOT00000046879
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr2_+_86996798 | 0.15 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr11_-_54567357 | 0.08 |
ENSRNOT00000046890
|
Retnlb
|
resistin like beta |
| chr20_+_8325041 | 0.05 |
ENSRNOT00000000640
|
Cmtr1
|
cap methyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 24.7 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 2.7 | 8.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 2.4 | 7.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 1.7 | 5.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.7 | 5.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 1.3 | 4.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.3 | 5.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 1.3 | 3.8 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.2 | 4.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.9 | 8.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.6 | 3.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 10.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.5 | 3.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 2.7 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 6.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.4 | 1.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 1.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 1.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.4 | 10.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.3 | 2.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.3 | 2.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 5.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 2.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 5.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 3.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 3.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 10.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 3.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 7.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 5.4 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 0.3 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.1 | 6.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 3.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.9 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 6.6 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 4.2 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.7 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 12.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 6.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.5 | 13.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 2.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.3 | 4.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 4.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 1.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 3.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 5.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 5.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 5.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 10.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 2.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 7.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 24.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 2.7 | 8.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 2.2 | 8.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.9 | 5.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.7 | 5.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.8 | 10.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 3.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 2.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 1.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 3.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 2.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 2.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 1.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 5.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 4.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 3.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 4.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 1.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 7.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 3.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 3.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 5.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 11.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 17.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 3.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 6.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 10.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 12.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 18.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 5.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 5.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 3.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 8.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 3.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 4.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 5.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |