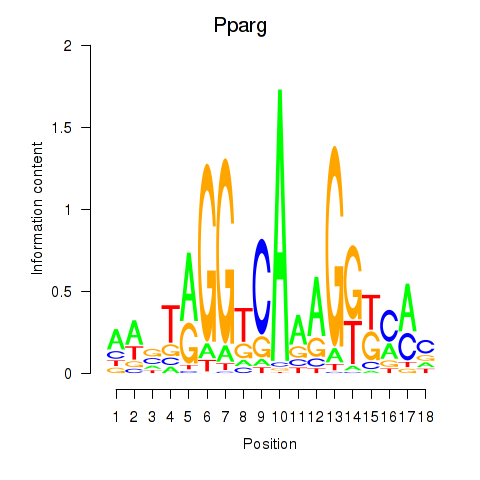
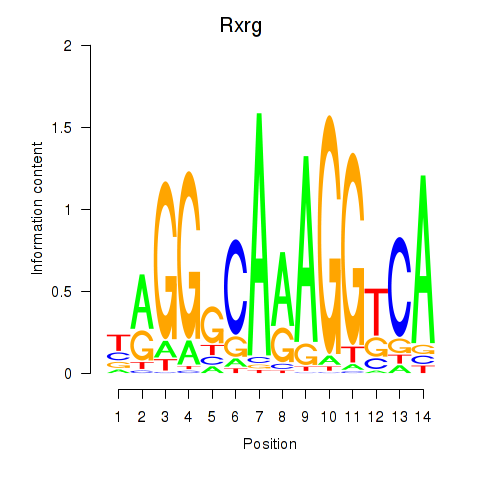
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pparg_Rxrg
Z-value: 3.01
Transcription factors associated with Pparg_Rxrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pparg
|
ENSRNOG00000008839 | peroxisome proliferator-activated receptor gamma |
|
Rxrg
|
ENSRNOG00000004537 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrg | rn6_v1_chr13_+_85818427_85818473 | 0.32 | 6.5e-09 | Click! |
| Pparg | rn6_v1_chr4_+_147275334_147275334 | -0.22 | 7.9e-05 | Click! |
Activity profile of Pparg_Rxrg motif
Sorted Z-values of Pparg_Rxrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_123638702 | 245.22 |
ENSRNOT00000082473
ENSRNOT00000044470 ENSRNOT00000092017 |
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr7_+_11490852 | 183.16 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr2_-_243407608 | 180.65 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr4_+_99063181 | 172.98 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr2_+_20857202 | 164.85 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr8_-_50531423 | 164.14 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr7_-_123621102 | 154.80 |
ENSRNOT00000046024
|
Cyp2d5
|
cytochrome P450, family 2, subfamily d, polypeptide 5 |
| chr14_+_22142364 | 154.74 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr9_+_16924520 | 152.52 |
ENSRNOT00000025094
|
Slc22a7
|
solute carrier family 22 member 7 |
| chr11_+_65022100 | 145.77 |
ENSRNOT00000003934
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr10_+_56662242 | 144.53 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_-_123655896 | 140.64 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr1_-_259287684 | 135.82 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr12_+_52452273 | 134.22 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chr7_-_123630045 | 133.21 |
ENSRNOT00000050002
|
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr10_+_56662561 | 131.50 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_-_30105132 | 128.02 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_-_56624526 | 124.93 |
ENSRNOT00000024973
|
Acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr2_-_178389608 | 122.52 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr1_-_258766881 | 122.17 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr16_-_81822716 | 117.70 |
ENSRNOT00000026677
|
F10
|
coagulation factor X |
| chr1_+_100470722 | 112.39 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr1_-_89543967 | 111.82 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr1_+_20856187 | 107.52 |
ENSRNOT00000071726
|
Smlr1
|
small leucine-rich protein 1 |
| chr2_+_188449210 | 104.91 |
ENSRNOT00000027700
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr18_+_70733872 | 104.87 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr16_-_81834945 | 103.86 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr2_+_200452624 | 103.62 |
ENSRNOT00000026121
|
Hmgcs2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 |
| chr3_+_171213936 | 100.45 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr1_+_88955440 | 95.98 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_+_219329574 | 95.14 |
ENSRNOT00000071109
|
Cabp2
|
calcium binding protein 2 |
| chr1_+_88955135 | 94.24 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chrX_-_13601069 | 94.11 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr2_+_60337667 | 92.84 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr20_-_12820466 | 91.15 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chrX_+_45420596 | 87.04 |
ENSRNOT00000051897
|
Sts
|
steroid sulfatase (microsomal), isozyme S |
| chr10_+_64952119 | 86.42 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr1_+_219833299 | 86.40 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr4_+_153876149 | 86.11 |
ENSRNOT00000018083
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr7_+_2689501 | 84.64 |
ENSRNOT00000041341
|
Apof
|
apolipoprotein F |
| chr1_+_83163079 | 84.35 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chrX_-_23139694 | 83.79 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr13_-_102721218 | 83.61 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr1_-_98486976 | 83.49 |
ENSRNOT00000024083
|
Etfb
|
electron transfer flavoprotein beta subunit |
| chr7_+_28412198 | 82.54 |
ENSRNOT00000081822
ENSRNOT00000038780 ENSRNOT00000005995 |
Igf1
|
insulin-like growth factor 1 |
| chr2_-_23256158 | 81.40 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr17_+_22620721 | 80.34 |
ENSRNOT00000019478
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr17_+_9736577 | 79.67 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr7_-_3386522 | 79.05 |
ENSRNOT00000010760
|
Mettl7b
|
methyltransferase like 7B |
| chr4_+_65110746 | 78.38 |
ENSRNOT00000017675
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr1_+_282567674 | 76.90 |
ENSRNOT00000090543
|
Ces2i
|
carboxylesterase 2I |
| chr14_+_77079402 | 75.88 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr13_+_56598957 | 75.23 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr7_-_70661891 | 74.23 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr13_+_89597138 | 73.54 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr17_+_9736786 | 73.34 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr8_-_60086403 | 72.62 |
ENSRNOT00000020544
|
Etfa
|
electron transfer flavoprotein alpha subunit |
| chr4_-_176679815 | 71.64 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr13_+_85580828 | 71.05 |
ENSRNOT00000005611
|
Aldh9a1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr10_-_38774449 | 70.94 |
ENSRNOT00000049820
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr18_-_24929091 | 70.63 |
ENSRNOT00000019596
|
Proc
|
protein C |
| chr3_+_8832740 | 69.85 |
ENSRNOT00000022552
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr3_-_14112851 | 69.72 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr11_-_81660395 | 69.62 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr8_-_130550388 | 67.98 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr10_-_103848035 | 67.65 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr12_-_48238887 | 66.34 |
ENSRNOT00000078868
|
Acacb
|
acetyl-CoA carboxylase beta |
| chr20_+_28989491 | 63.82 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr11_-_81639872 | 62.63 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chr5_+_165415136 | 62.24 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr8_-_119265157 | 61.69 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr5_+_120250616 | 61.52 |
ENSRNOT00000070967
|
Ak4
|
adenylate kinase 4 |
| chr1_-_198486157 | 61.36 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr20_-_4561062 | 60.15 |
ENSRNOT00000065044
ENSRNOT00000092698 ENSRNOT00000060607 |
Cfb
C2
|
complement factor B complement C2 |
| chr8_-_111777602 | 59.93 |
ENSRNOT00000052317
ENSRNOT00000083855 |
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr2_+_114413410 | 58.62 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr1_+_213511874 | 58.60 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr2_-_53313884 | 58.55 |
ENSRNOT00000046951
|
Ghr
|
growth hormone receptor |
| chr1_-_224533219 | 57.42 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr1_+_266482858 | 56.76 |
ENSRNOT00000027223
|
As3mt
|
arsenite methyltransferase |
| chr2_-_260148589 | 56.18 |
ENSRNOT00000013238
|
Acadm
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr9_-_61690956 | 55.93 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr18_-_35071619 | 55.33 |
ENSRNOT00000075695
|
LOC100911558
|
serine protease inhibitor Kazal-type 3-like |
| chr18_-_38185812 | 54.93 |
ENSRNOT00000017969
|
Spink1l
|
serine peptidase inhibitor, Kazal type 1-like |
| chr12_+_47254484 | 54.73 |
ENSRNOT00000001556
|
Acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chrX_+_21696772 | 54.31 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr7_+_3216497 | 54.16 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr4_+_100218661 | 53.34 |
ENSRNOT00000079415
|
Tmem150a
|
transmembrane protein 150A |
| chr1_-_146736261 | 53.17 |
ENSRNOT00000068167
|
Fah
|
fumarylacetoacetate hydrolase |
| chr1_-_89369960 | 52.93 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr1_+_100471066 | 52.55 |
ENSRNOT00000067562
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr1_-_170431073 | 52.42 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr10_+_96639924 | 51.86 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr18_-_36285444 | 51.59 |
ENSRNOT00000087852
|
Prelid2
|
PRELI domain containing 2 |
| chr1_-_148119857 | 51.55 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr16_+_54153054 | 51.50 |
ENSRNOT00000090644
ENSRNOT00000014248 |
Fgl1
|
fibrinogen-like 1 |
| chr10_+_109708755 | 51.44 |
ENSRNOT00000083601
|
Gcgr
|
glucagon receptor |
| chr9_-_4978892 | 50.61 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr2_+_84645084 | 50.47 |
ENSRNOT00000015448
|
Cmbl
|
carboxymethylenebutenolidase homolog |
| chr3_-_51054378 | 48.90 |
ENSRNOT00000089243
|
Grb14
|
growth factor receptor bound protein 14 |
| chr12_-_48218955 | 48.09 |
ENSRNOT00000067975
ENSRNOT00000080557 ENSRNOT00000000821 |
Acacb
|
acetyl-CoA carboxylase beta |
| chr3_+_114355798 | 47.88 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chr7_-_143793774 | 47.73 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr10_+_86399827 | 47.60 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr5_+_117698764 | 47.23 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr7_-_143793970 | 46.29 |
ENSRNOT00000016205
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr1_+_193424812 | 46.18 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chr2_+_243701962 | 46.14 |
ENSRNOT00000016891
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr16_+_18736154 | 46.10 |
ENSRNOT00000015723
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr4_-_176381477 | 45.45 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr7_-_14435967 | 44.62 |
ENSRNOT00000074801
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr4_-_176294997 | 44.59 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr1_+_264741911 | 44.56 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr14_-_84334066 | 44.50 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr13_-_85622314 | 44.36 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr3_+_154786215 | 43.72 |
ENSRNOT00000019787
|
Lbp
|
lipopolysaccharide binding protein |
| chr13_+_82496022 | 43.65 |
ENSRNOT00000080759
|
F5
|
coagulation factor V |
| chr2_+_219563783 | 43.63 |
ENSRNOT00000020267
|
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr10_-_109909646 | 43.62 |
ENSRNOT00000074362
ENSRNOT00000088907 |
Dcxr
|
dicarbonyl and L-xylulose reductase |
| chr1_+_189514553 | 43.58 |
ENSRNOT00000020039
|
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_+_282694906 | 43.46 |
ENSRNOT00000074303
|
Ces2c
|
carboxylesterase 2C |
| chr6_+_107460668 | 41.74 |
ENSRNOT00000013515
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr2_+_60949256 | 41.73 |
ENSRNOT00000025323
ENSRNOT00000040701 |
Amacr
|
alpha-methylacyl-CoA racemase |
| chr2_+_221823687 | 41.72 |
ENSRNOT00000072735
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr2_+_243502073 | 41.58 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr3_-_150108898 | 41.41 |
ENSRNOT00000022914
|
Pxmp4
|
peroxisomal membrane protein 4 |
| chr19_+_487723 | 40.87 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr8_+_117455262 | 40.26 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr17_+_69468427 | 40.18 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr20_+_13827132 | 39.92 |
ENSRNOT00000001664
|
Ddt
|
D-dopachrome tautomerase |
| chr7_-_117187367 | 39.24 |
ENSRNOT00000014191
|
LOC680875
|
similar to dystonin isoform 1 |
| chr20_+_14101659 | 39.16 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr9_+_95295701 | 38.25 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr17_-_43543172 | 37.89 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr7_+_71057911 | 37.66 |
ENSRNOT00000037218
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr10_+_103396155 | 37.57 |
ENSRNOT00000086924
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr8_-_77398156 | 37.49 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr3_-_5481144 | 37.09 |
ENSRNOT00000078429
|
Surf4
|
surfeit 4 |
| chr3_-_7141522 | 37.08 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr10_-_70802782 | 36.60 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr2_+_243656579 | 36.31 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr10_+_90085559 | 36.21 |
ENSRNOT00000028332
|
Nags
|
N-acetylglutamate synthase |
| chr12_-_41627741 | 36.18 |
ENSRNOT00000001875
|
Sds
|
serine dehydratase |
| chr13_-_50549981 | 35.95 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr1_+_221236773 | 35.89 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr1_-_212579057 | 35.79 |
ENSRNOT00000025446
|
LOC100911186
|
enoyl-CoA hydratase, mitochondrial-like |
| chr9_+_4807717 | 35.75 |
ENSRNOT00000092159
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr17_+_22619891 | 35.66 |
ENSRNOT00000060403
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr6_-_10899200 | 35.52 |
ENSRNOT00000089104
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr5_+_76812931 | 35.35 |
ENSRNOT00000059458
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr2_+_235738416 | 35.14 |
ENSRNOT00000074209
|
Etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr1_-_261853495 | 34.44 |
ENSRNOT00000021144
|
Loxl4
|
lysyl oxidase-like 4 |
| chr8_+_103342929 | 34.43 |
ENSRNOT00000045455
|
LOC501038
|
Ab2-060 |
| chr18_+_46148849 | 34.27 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr1_-_258875572 | 34.17 |
ENSRNOT00000093005
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr14_+_17195014 | 33.83 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr14_-_86164341 | 33.45 |
ENSRNOT00000086343
ENSRNOT00000080147 |
Gck
|
glucokinase |
| chr12_-_48627297 | 32.84 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr10_-_62254287 | 32.69 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr9_+_4269160 | 32.44 |
ENSRNOT00000050420
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr6_+_26051396 | 32.23 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr10_+_103395511 | 32.01 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr9_+_81689802 | 31.93 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr2_+_198965685 | 31.76 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr12_+_30441055 | 31.75 |
ENSRNOT00000081099
|
AABR07036007.1
|
|
| chr3_-_93412058 | 31.72 |
ENSRNOT00000011230
|
Cat
|
catalase |
| chr1_-_226049929 | 31.00 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr2_-_148722263 | 30.78 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr20_-_45053640 | 30.68 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr6_-_26385761 | 30.49 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr7_+_143754892 | 30.46 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr1_-_222178725 | 30.37 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr7_+_140781799 | 30.36 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr10_+_71202456 | 30.18 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr19_+_15139920 | 30.04 |
ENSRNOT00000074479
ENSRNOT00000020775 |
Ces1f
|
carboxylesterase 1F |
| chr20_-_3819200 | 29.90 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_198559568 | 29.86 |
ENSRNOT00000023080
|
Qprt
|
quinolinate phosphoribosyltransferase |
| chr4_+_14109864 | 29.74 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr5_-_57372239 | 29.61 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr7_+_12314848 | 29.57 |
ENSRNOT00000028969
|
Gamt
|
guanidinoacetate N-methyltransferase |
| chr9_+_11064605 | 29.42 |
ENSRNOT00000075121
|
Stap2
|
signal transducing adaptor family member 2 |
| chr4_-_145390447 | 28.92 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr20_+_13827660 | 28.70 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr7_-_12246729 | 28.61 |
ENSRNOT00000044030
|
Reep6
|
receptor accessory protein 6 |
| chr2_-_210809306 | 28.51 |
ENSRNOT00000047139
|
Gstm1
|
glutathione S-transferase mu 1 |
| chr18_+_44810388 | 28.29 |
ENSRNOT00000021646
ENSRNOT00000089750 |
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr2_+_227455722 | 28.27 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr1_+_213686046 | 27.95 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr13_-_112005052 | 27.56 |
ENSRNOT00000007879
|
G0s2
|
G0/G1switch 2 |
| chr1_-_53038229 | 27.38 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr5_+_78222504 | 27.24 |
ENSRNOT00000019544
|
Slc31a1
|
solute carrier family 31 member 1 |
| chr3_-_46361092 | 27.04 |
ENSRNOT00000008987
|
Cd302
|
CD302 molecule |
| chr13_-_80703615 | 26.94 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr8_+_91464229 | 26.78 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr1_+_35067490 | 25.97 |
ENSRNOT00000022790
|
Adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr2_+_188449718 | 25.83 |
ENSRNOT00000065791
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr14_-_79464770 | 25.72 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr11_+_87204175 | 25.62 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr2_+_56426367 | 25.57 |
ENSRNOT00000016036
|
Lifr
|
leukemia inhibitory factor receptor alpha |
| chr20_-_4530126 | 25.28 |
ENSRNOT00000000481
|
Skiv2l
|
Ski2 like RNA helicase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pparg_Rxrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 79.2 | 237.7 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 63.4 | 190.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 57.7 | 173.0 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 55.0 | 164.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 51.0 | 153.0 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 50.7 | 608.6 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 46.8 | 514.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 42.7 | 128.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 40.7 | 81.4 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 37.3 | 111.8 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 34.8 | 104.5 | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 31.3 | 94.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 30.9 | 92.8 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 30.4 | 91.2 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 29.3 | 58.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 29.1 | 232.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 28.6 | 114.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 27.9 | 83.6 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 26.1 | 130.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 25.9 | 103.6 | GO:0034696 | response to prostaglandin F(GO:0034696) ketone body biosynthetic process(GO:0046951) |
| 25.1 | 100.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 23.6 | 94.3 | GO:0019477 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 22.1 | 154.7 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 21.6 | 86.4 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 21.0 | 104.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 20.9 | 62.6 | GO:0097037 | heme export(GO:0097037) |
| 20.9 | 166.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 20.7 | 124.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 20.6 | 82.5 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 18.6 | 55.9 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 18.4 | 18.4 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 18.1 | 72.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 18.0 | 72.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 16.6 | 49.8 | GO:0061215 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 16.2 | 372.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 14.6 | 43.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 14.5 | 43.6 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 14.3 | 114.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 14.2 | 71.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 13.9 | 41.7 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 13.6 | 204.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 13.5 | 148.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 13.2 | 52.9 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 13.1 | 52.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 12.5 | 75.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 12.5 | 37.5 | GO:0010034 | response to acetate(GO:0010034) |
| 12.5 | 25.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 12.2 | 183.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 11.7 | 23.5 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 11.7 | 58.6 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 11.1 | 44.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 10.7 | 32.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 10.2 | 40.9 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 9.8 | 88.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 9.8 | 48.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 9.6 | 115.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 9.6 | 76.4 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 9.5 | 28.5 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 9.1 | 27.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 8.9 | 44.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 8.8 | 309.4 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 8.4 | 33.5 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 8.3 | 24.9 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 8.0 | 47.9 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 7.7 | 108.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 7.6 | 197.6 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 7.4 | 22.3 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 7.3 | 43.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 7.2 | 35.9 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 7.2 | 43.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 7.1 | 113.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 6.8 | 27.2 | GO:1902861 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 6.7 | 40.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 6.6 | 46.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 6.4 | 51.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 6.4 | 25.6 | GO:0015746 | citrate transport(GO:0015746) |
| 6.4 | 25.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 6.4 | 31.9 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 6.1 | 18.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 6.0 | 29.9 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 6.0 | 41.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 5.9 | 11.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 5.8 | 110.3 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 5.8 | 23.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 5.6 | 16.8 | GO:1902988 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 5.5 | 38.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 5.4 | 26.9 | GO:0042737 | drug catabolic process(GO:0042737) |
| 5.4 | 16.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) pulmonary vein morphogenesis(GO:0060577) |
| 5.4 | 37.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 5.3 | 21.3 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 5.1 | 15.4 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 5.1 | 15.3 | GO:0003032 | age-dependent response to oxidative stress(GO:0001306) detection of oxygen(GO:0003032) age-dependent general metabolic decline(GO:0007571) |
| 5.1 | 15.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 5.1 | 40.5 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 5.0 | 25.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 5.0 | 125.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 5.0 | 24.9 | GO:0070541 | response to platinum ion(GO:0070541) |
| 4.8 | 23.9 | GO:0060112 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 4.8 | 14.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 4.7 | 23.7 | GO:0060744 | positive regulation of exit from mitosis(GO:0031536) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 4.7 | 18.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 4.7 | 14.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 4.7 | 14.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 4.7 | 9.4 | GO:0043465 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 4.7 | 32.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 4.5 | 13.5 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 4.4 | 13.3 | GO:1901492 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 4.4 | 13.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 4.3 | 13.0 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 4.3 | 21.5 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 4.3 | 21.4 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 4.3 | 38.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 4.2 | 12.7 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 4.2 | 12.7 | GO:2000360 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 4.2 | 20.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 4.0 | 43.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 4.0 | 31.7 | GO:0010193 | response to ozone(GO:0010193) |
| 3.9 | 27.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 3.9 | 15.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 3.9 | 11.6 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 3.8 | 7.6 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 3.8 | 18.8 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 3.7 | 47.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 3.6 | 14.4 | GO:0033030 | negative regulation of hypersensitivity(GO:0002884) negative regulation of neutrophil apoptotic process(GO:0033030) |
| 3.6 | 14.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 3.6 | 14.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 3.5 | 56.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 3.5 | 38.8 | GO:0001554 | luteolysis(GO:0001554) |
| 3.5 | 17.6 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 3.5 | 10.4 | GO:0030091 | protein repair(GO:0030091) |
| 3.5 | 10.4 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 3.4 | 6.8 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 3.4 | 3.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.4 | 10.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 3.3 | 10.0 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 3.3 | 6.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 3.2 | 41.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 3.2 | 9.6 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 3.1 | 59.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 3.1 | 18.7 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 3.1 | 9.3 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 3.1 | 61.7 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 3.1 | 9.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.0 | 15.2 | GO:0015747 | urate transport(GO:0015747) |
| 2.9 | 8.8 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) protein localization to site of double-strand break(GO:1990166) |
| 2.9 | 40.8 | GO:0051923 | sulfation(GO:0051923) |
| 2.8 | 19.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 2.8 | 39.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 2.8 | 25.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 2.8 | 14.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 2.8 | 8.3 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 2.8 | 85.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 2.8 | 38.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 2.7 | 49.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 2.7 | 54.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 2.7 | 24.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 2.7 | 8.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 2.7 | 2.7 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 2.6 | 13.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 2.6 | 10.6 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 2.6 | 10.5 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 2.6 | 5.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 2.5 | 37.9 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 2.5 | 10.1 | GO:0009597 | detection of virus(GO:0009597) |
| 2.5 | 10.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.5 | 17.6 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 2.5 | 12.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) ether lipid metabolic process(GO:0046485) |
| 2.4 | 26.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 2.4 | 9.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 2.4 | 28.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 2.3 | 9.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 2.3 | 11.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 2.3 | 20.9 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 2.3 | 39.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 2.3 | 22.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.3 | 11.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 2.3 | 11.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 2.2 | 8.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 2.2 | 15.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 2.2 | 17.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.2 | 32.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 2.1 | 38.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 2.1 | 33.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 2.1 | 6.3 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 2.0 | 8.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 2.0 | 12.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 2.0 | 16.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.0 | 5.9 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 2.0 | 11.9 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.9 | 34.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.8 | 14.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.8 | 78.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 1.8 | 5.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.8 | 9.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.7 | 8.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.7 | 8.7 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.7 | 49.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 1.7 | 8.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.7 | 42.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.7 | 8.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.7 | 11.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.7 | 9.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 1.6 | 32.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 1.6 | 6.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) endodermal digestive tract morphogenesis(GO:0061031) |
| 1.6 | 6.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 1.6 | 52.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 1.6 | 7.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.6 | 18.8 | GO:0009750 | response to fructose(GO:0009750) |
| 1.6 | 12.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.5 | 60.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 1.5 | 9.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) response to amino acid starvation(GO:1990928) |
| 1.5 | 3.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.5 | 3.0 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 1.5 | 23.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 1.5 | 8.8 | GO:0042148 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 1.4 | 21.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.4 | 8.6 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 1.4 | 7.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.4 | 2.9 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.4 | 10.0 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 1.4 | 30.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 1.4 | 15.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.4 | 5.6 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 1.4 | 16.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.4 | 4.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 1.3 | 4.0 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 1.3 | 1.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.3 | 12.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 1.3 | 8.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.3 | 9.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.3 | 10.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 1.3 | 51.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 1.3 | 36.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 1.3 | 14.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.3 | 3.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.3 | 2.6 | GO:0061724 | lipophagy(GO:0061724) |
| 1.3 | 2.6 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 1.3 | 20.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 1.3 | 2.5 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 1.3 | 25.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 1.3 | 5.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.3 | 3.8 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 1.2 | 8.7 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 1.2 | 9.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 1.2 | 57.8 | GO:0009268 | response to pH(GO:0009268) |
| 1.2 | 7.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.2 | 46.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 1.2 | 3.5 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) RNA 5'-end processing(GO:0000966) RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) polyadenylation-dependent RNA catabolic process(GO:0043633) |
| 1.2 | 12.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.2 | 22.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 1.1 | 2.3 | GO:0009992 | primary ovarian follicle growth(GO:0001545) cellular water homeostasis(GO:0009992) |
| 1.1 | 11.4 | GO:1902003 | regulation of beta-amyloid formation(GO:1902003) |
| 1.1 | 3.4 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 4.5 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.1 | 15.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 1.1 | 6.6 | GO:0051503 | ADP transport(GO:0015866) ATP transport(GO:0015867) adenine nucleotide transport(GO:0051503) |
| 1.1 | 19.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.1 | 10.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.1 | 3.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.1 | 4.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 1.1 | 7.4 | GO:0060613 | fat pad development(GO:0060613) |
| 1.0 | 12.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.0 | 30.8 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 1.0 | 3.0 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 13.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 1.0 | 171.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 1.0 | 12.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.0 | 3.9 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 1.0 | 25.3 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 1.0 | 7.7 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.9 | 10.4 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.9 | 36.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.9 | 7.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.9 | 8.5 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.9 | 20.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.9 | 3.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.9 | 18.3 | GO:0019915 | lipid storage(GO:0019915) |
| 0.9 | 10.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.9 | 2.7 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.9 | 3.5 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.9 | 8.7 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.9 | 7.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.9 | 27.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.8 | 20.9 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.8 | 2.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.8 | 15.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.8 | 174.1 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.8 | 15.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.8 | 51.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.8 | 20.8 | GO:0006956 | complement activation(GO:0006956) |
| 0.8 | 4.8 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.8 | 7.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.8 | 5.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.8 | 3.9 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.8 | 39.8 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.8 | 8.5 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.8 | 1.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 13.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.7 | 12.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.7 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.7 | 14.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.7 | 23.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.7 | 15.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.7 | 7.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.7 | 14.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.7 | 11.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.7 | 31.4 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.7 | 7.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.7 | 5.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.7 | 5.9 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.7 | 3.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.7 | 14.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.6 | 2.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.6 | 11.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.6 | 6.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.6 | 26.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.6 | 15.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.6 | 22.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.6 | 8.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.6 | 4.2 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.6 | 48.5 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.6 | 4.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.6 | 1.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 14.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.5 | 6.0 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.5 | 9.0 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.5 | 1.6 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.5 | 9.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.5 | 6.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.5 | 1.4 | GO:0061043 | positive regulation of vascular wound healing(GO:0035470) regulation of vascular wound healing(GO:0061043) |
| 0.5 | 3.7 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.5 | 4.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 18.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 21.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.4 | 3.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.4 | 6.2 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.4 | 2.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 11.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 3.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 3.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 5.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 5.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.4 | 1.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.3 | 2.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.3 | 1.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.3 | 6.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.3 | 2.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 3.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.3 | 1.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 2.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 1.1 | GO:1902416 | regulation of mRNA binding(GO:1902415) positive regulation of mRNA binding(GO:1902416) |
| 0.3 | 5.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 2.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 4.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 5.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 10.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.2 | 1.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.5 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.2 | 2.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 2.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 7.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 4.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.2 | 4.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.2 | 92.3 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.2 | 4.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 0.6 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 0.5 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.4 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 15.7 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.1 | 0.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.6 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.1 | 1.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 126.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.9 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 3.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 5.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.5 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 5.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 3.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 1.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 1.9 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.6 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 28.3 | 254.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 22.6 | 225.9 | GO:0045179 | apical cortex(GO:0045179) |
| 18.2 | 91.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 16.5 | 82.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 15.7 | 62.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 15.4 | 46.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 11.7 | 58.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 11.2 | 55.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 10.0 | 70.2 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 10.0 | 69.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 9.6 | 182.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 9.5 | 28.6 | GO:0044317 | rod spherule(GO:0044317) |
| 8.7 | 174.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 8.4 | 25.3 | GO:0055087 | Ski complex(GO:0055087) |
| 8.0 | 199.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 6.1 | 24.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 6.0 | 18.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 5.4 | 167.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 5.4 | 26.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 4.9 | 256.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 4.7 | 32.7 | GO:0043203 | axon hillock(GO:0043203) |
| 4.6 | 27.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 4.5 | 13.5 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) |
| 4.4 | 140.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 4.3 | 21.5 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 4.0 | 12.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 3.9 | 1056.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 3.8 | 11.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 3.6 | 14.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 3.5 | 3.5 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 3.5 | 51.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 3.2 | 9.7 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 3.1 | 9.4 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 2.8 | 38.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 2.7 | 8.1 | GO:0042627 | chylomicron(GO:0042627) |
| 2.6 | 7.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 2.5 | 7.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 2.5 | 5.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 2.3 | 96.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 2.3 | 6.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 2.2 | 6.6 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 2.1 | 31.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 2.1 | 12.7 | GO:0097422 | tubular endosome(GO:0097422) |
| 2.0 | 1643.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 2.0 | 626.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 1.8 | 38.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 1.8 | 12.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.8 | 8.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.6 | 42.0 | GO:0005902 | microvillus(GO:0005902) |
| 1.5 | 160.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.5 | 124.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 1.5 | 17.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.5 | 13.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.4 | 8.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.4 | 73.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 1.4 | 4.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.3 | 3.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.3 | 20.9 | GO:0005883 | neurofilament(GO:0005883) |
| 1.2 | 19.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.2 | 11.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.2 | 10.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.2 | 6.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.1 | 4.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.1 | 57.0 | GO:0005811 | lipid particle(GO:0005811) |
| 1.1 | 63.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 1.0 | 7.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.0 | 50.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 1.0 | 140.2 | GO:0005903 | brush border(GO:0005903) |
| 0.9 | 8.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.9 | 11.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.9 | 14.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.9 | 11.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.9 | 9.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.9 | 4.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 110.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.8 | 4.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.8 | 2.4 | GO:0000813 | ESCRT I complex(GO:0000813) ESCRT complex(GO:0036452) |
| 0.7 | 14.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.7 | 6.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.7 | 8.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 3.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.6 | 21.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.6 | 38.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.6 | 659.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.6 | 7.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.6 | 8.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.6 | 8.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 83.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.5 | 115.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.5 | 22.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.5 | 6.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.5 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 14.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 13.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 2.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 4.9 | GO:0030057 | desmosome(GO:0030057) messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 24.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.3 | 8.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 5.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.3 | 372.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 2.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 3.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 10.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 35.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 60.6 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.2 | 10.5 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 43.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.2 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 2.8 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 2.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 3.6 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.2 | 55.3 | GO:0031967 | organelle envelope(GO:0031967) |
| 0.2 | 2.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 0.2 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 0.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 5.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 1.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 215.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 79.2 | 237.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 63.4 | 190.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 43.9 | 175.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 41.6 | 124.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 33.4 | 301.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 32.7 | 130.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 29.3 | 205.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 28.8 | 86.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 28.6 | 114.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 26.1 | 78.4 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 25.6 | 128.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 25.5 | 1047.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 25.5 | 127.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 23.9 | 143.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 23.5 | 70.6 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 23.2 | 69.7 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 22.9 | 68.6 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 21.6 | 86.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 20.9 | 83.6 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 20.7 | 124.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 20.4 | 163.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 18.9 | 94.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 18.4 | 18.4 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 17.7 | 53.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 17.3 | 51.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 16.5 | 82.6 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 16.3 | 114.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 15.0 | 104.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 14.6 | 58.6 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 14.4 | 43.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 14.4 | 86.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 13.9 | 292.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 13.9 | 41.7 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 13.4 | 40.3 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 12.9 | 129.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 12.9 | 51.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 12.4 | 37.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 12.3 | 61.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 12.0 | 47.9 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 11.9 | 190.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 11.1 | 44.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 11.1 | 155.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 10.9 | 32.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 10.9 | 43.5 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 10.6 | 53.1 | GO:0070404 | NADH binding(GO:0070404) |
| 10.6 | 31.7 | GO:0004046 | aminoacylase activity(GO:0004046) catalase activity(GO:0004096) |
| 10.2 | 71.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 10.2 | 40.9 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 10.2 | 81.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 9.9 | 119.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 9.9 | 29.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 9.1 | 27.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 9.1 | 36.4 | GO:0034618 | arginine binding(GO:0034618) |
| 8.7 | 43.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 8.5 | 110.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 8.5 | 25.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 8.4 | 167.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 8.2 | 115.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 8.0 | 31.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 7.7 | 15.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 7.5 | 29.9 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 7.4 | 14.9 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 7.4 | 22.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 7.4 | 29.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 7.3 | 36.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 6.9 | 34.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 6.8 | 33.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 6.6 | 52.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 6.5 | 58.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 6.5 | 155.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 6.4 | 25.6 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 6.1 | 30.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 6.0 | 12.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 6.0 | 83.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 5.9 | 35.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 5.7 | 62.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 5.6 | 33.5 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 5.5 | 27.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 5.4 | 43.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 5.4 | 43.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 5.4 | 21.5 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 5.3 | 21.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 5.3 | 16.0 | GO:0004040 | amidase activity(GO:0004040) |
| 5.2 | 47.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 4.9 | 68.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 4.9 | 77.6 | GO:0043176 | amine binding(GO:0043176) |
| 4.8 | 43.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 4.8 | 105.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 4.8 | 14.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 4.6 | 13.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 4.6 | 27.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 4.6 | 13.8 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 4.5 | 49.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 4.5 | 18.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 4.5 | 35.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 4.5 | 13.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 4.5 | 13.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 4.4 | 17.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 4.4 | 39.4 | GO:0015250 | water channel activity(GO:0015250) |
| 4.3 | 17.1 | GO:0016160 | amylase activity(GO:0016160) |
| 4.0 | 271.1 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 4.0 | 20.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 3.7 | 22.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 3.6 | 28.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 3.4 | 27.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 3.4 | 57.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 3.4 | 23.7 | GO:0008430 | selenium binding(GO:0008430) |
| 3.2 | 12.8 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 3.1 | 15.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.0 | 39.6 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 3.0 | 26.8 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 2.9 | 17.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 2.9 | 177.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 2.9 | 17.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 2.8 | 22.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.8 | 11.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 2.8 | 19.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 2.8 | 67.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 2.8 | 14.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 2.8 | 13.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 2.8 | 41.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 2.7 | 8.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 2.7 | 541.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 2.7 | 10.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 2.7 | 8.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 2.6 | 10.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 2.6 | 10.4 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 2.6 | 10.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 2.6 | 25.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 2.5 | 20.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 2.5 | 37.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 2.5 | 14.9 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 2.5 | 22.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 2.5 | 14.8 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 2.4 | 16.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 2.3 | 91.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 2.3 | 41.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 2.3 | 27.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.3 | 9.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.2 | 15.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 2.2 | 17.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 2.2 | 68.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 2.2 | 33.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 2.1 | 99.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 2.0 | 124.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 2.0 | 62.5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 1.9 | 9.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.9 | 9.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.9 | 9.4 | GO:0070728 | leucine binding(GO:0070728) |
| 1.8 | 14.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.7 | 5.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 1.7 | 32.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 1.7 | 8.4 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 1.6 | 13.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.6 | 6.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.6 | 37.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 1.6 | 11.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.6 | 27.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 1.6 | 11.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.5 | 4.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 1.5 | 8.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.4 | 33.3 | GO:0070001 | aspartic-type peptidase activity(GO:0070001) |
| 1.4 | 4.2 | GO:0070698 | nodal binding(GO:0038100) type I activin receptor binding(GO:0070698) |
| 1.3 | 5.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.3 | 22.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 1.3 | 3.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.3 | 8.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.2 | 18.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.2 | 38.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.2 | 22.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.2 | 10.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.1 | 122.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.1 | 8.4 | GO:0071253 | connexin binding(GO:0071253) |
| 1.0 | 3.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.0 | 11.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.0 | 12.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 1.0 | 11.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.0 | 7.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 1.0 | 49.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 1.0 | 6.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.0 | 13.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.0 | 20.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.9 | 4.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.9 | 13.1 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.9 | 13.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.9 | 17.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.9 | 17.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 5.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.9 | 4.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.9 | 5.4 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) |
| 0.9 | 2.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.9 | 10.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.8 | 11.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.8 | 52.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.8 | 4.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.8 | 3.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.8 | 147.3 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.8 | 6.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.8 | 19.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.7 | 10.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.7 | 6.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.7 | 7.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.7 | 1.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.7 | 17.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.7 | 5.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.7 | 11.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.7 | 9.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 14.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.7 | 7.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.6 | 22.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.6 | 32.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.6 | 47.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.6 | 3.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.6 | 14.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 2.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.6 | 3.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.6 | 11.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.6 | 4.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.5 | 63.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.5 | 2.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 10.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.5 | 1.5 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.5 | 55.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 8.6 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.4 | 2.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 2.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 9.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 18.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.4 | 1.6 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 7.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.4 | 14.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.4 | 2.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 7.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.4 | 2.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.4 | 4.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 5.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 10.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.3 | 5.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 11.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.3 | 1.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 21.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 9.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 8.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 4.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 2.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 5.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 5.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 3.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 6.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 11.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.2 | 2.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 73.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 0.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 4.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 8.8 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 9.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.6 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 5.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 7.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 51.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 581.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 5.3 | 163.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 4.7 | 121.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 3.1 | 134.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 2.7 | 98.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 2.6 | 62.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 2.5 | 599.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 2.4 | 112.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 2.2 | 4.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 2.1 | 159.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 1.8 | 55.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 1.8 | 35.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 1.3 | 15.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.2 | 43.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 1.1 | 18.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 1.0 | 49.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.9 | 30.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.9 | 20.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.8 | 8.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.8 | 28.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.6 | 11.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.6 | 9.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.6 | 9.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.4 | 25.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.4 | 16.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 14.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.3 | 5.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 9.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 6.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 10.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 3.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 15.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 2.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 12.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 13.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 3.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 7.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.2 | 292.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 27.3 | 463.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 19.5 | 58.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 19.5 | 273.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 16.8 | 101.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 15.7 | 188.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 14.6 | 160.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 14.4 | 187.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 11.0 | 87.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 10.4 | 134.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 9.9 | 118.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 8.7 | 8.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 8.7 | 87.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 8.5 | 153.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 8.4 | 50.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 8.3 | 149.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 8.1 | 137.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 7.1 | 205.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 7.1 | 141.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 7.1 | 155.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 7.0 | 83.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 6.8 | 109.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 5.9 | 229.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 5.4 | 353.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 5.4 | 32.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 5.4 | 112.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 5.0 | 69.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 4.8 | 100.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 3.9 | 38.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 3.8 | 65.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 3.8 | 101.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 3.2 | 371.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 3.2 | 35.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 2.9 | 17.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 2.6 | 18.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 2.6 | 389.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 2.5 | 152.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 2.3 | 63.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 2.3 | 38.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.2 | 30.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 2.1 | 63.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 2.0 | 32.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.8 | 74.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.8 | 58.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 1.7 | 13.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.6 | 19.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.3 | 2.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 1.3 | 53.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 1.3 | 20.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 1.2 | 61.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 1.2 | 23.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.2 | 16.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.2 | 40.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 1.1 | 17.9 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 1.1 | 7.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.1 | 20.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 1.0 | 9.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.0 | 10.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 1.0 | 12.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.0 | 38.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.9 | 6.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.9 | 18.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.8 | 31.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.7 | 8.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 8.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.6 | 12.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.6 | 9.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.6 | 7.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 8.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 13.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 13.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.5 | 9.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 11.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 5.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 5.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 2.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 11.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 6.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 3.1 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.2 | 10.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 4.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 4.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 5.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |