Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
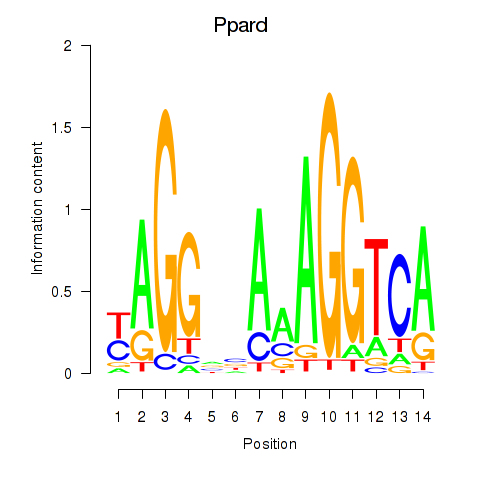
Results for Ppard
Z-value: 0.49
Transcription factors associated with Ppard
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppard
|
ENSRNOG00000000503 | peroxisome proliferator-activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppard | rn6_v1_chr20_+_7821755_7821755 | -0.31 | 1.1e-08 | Click! |
Activity profile of Ppard motif
Sorted Z-values of Ppard motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_57372239 | 32.38 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr2_-_178389608 | 21.69 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr12_-_48238887 | 21.28 |
ENSRNOT00000078868
|
Acacb
|
acetyl-CoA carboxylase beta |
| chr1_+_48273611 | 17.04 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr5_-_169301728 | 15.08 |
ENSRNOT00000013646
|
Espn
|
espin |
| chr7_+_11490852 | 14.34 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr14_+_22142364 | 12.69 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr2_+_198965685 | 11.23 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr14_-_79464770 | 10.86 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr8_+_117455262 | 10.20 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr10_-_56624526 | 9.49 |
ENSRNOT00000024973
|
Acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr12_+_47254484 | 9.11 |
ENSRNOT00000001556
|
Acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr5_+_76812931 | 8.45 |
ENSRNOT00000059458
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr20_+_14101659 | 6.68 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr2_+_200452624 | 6.56 |
ENSRNOT00000026121
|
Hmgcs2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 |
| chr14_-_84937725 | 6.42 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr10_+_71159869 | 6.12 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr11_-_24294179 | 5.23 |
ENSRNOT00000002116
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_+_26051396 | 5.07 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr1_-_222178725 | 4.94 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr5_+_165415136 | 4.75 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_+_95045034 | 4.74 |
ENSRNOT00000081305
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr8_+_117170620 | 4.56 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr20_-_3819200 | 4.40 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_153096269 | 4.32 |
ENSRNOT00000022578
|
Tmem135
|
transmembrane protein 135 |
| chr1_+_255842783 | 4.29 |
ENSRNOT00000023562
|
March5
|
membrane associated ring-CH-type finger 5 |
| chr3_-_60813869 | 4.00 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr10_+_31508512 | 3.98 |
ENSRNOT00000031580
|
Fam71b
|
family with sequence similarity 71, member B |
| chr10_+_84871680 | 3.95 |
ENSRNOT00000073292
|
Prr15l
|
proline rich 15-like |
| chr1_+_212558257 | 3.31 |
ENSRNOT00000024912
|
Prap1
|
proline-rich acidic protein 1 |
| chr7_-_143793774 | 3.09 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr2_+_244521699 | 2.81 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr9_-_69953182 | 2.75 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr7_-_143793970 | 2.60 |
ENSRNOT00000016205
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr3_+_81287242 | 2.54 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr3_-_120106697 | 2.47 |
ENSRNOT00000020354
|
Prom2
|
prominin 2 |
| chr10_+_11146359 | 2.44 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr9_+_64095978 | 2.34 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr10_-_88107232 | 2.13 |
ENSRNOT00000019330
ENSRNOT00000076770 |
Krt9
|
keratin 9 |
| chr8_-_119265157 | 1.99 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr13_+_96219853 | 1.68 |
ENSRNOT00000006154
|
Cox20
|
COX20 cytochrome C oxidase assembly factor |
| chr8_+_59164572 | 1.63 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr6_+_108363485 | 1.16 |
ENSRNOT00000073962
|
Vrtn
|
vertebrae development associated |
| chr5_-_167244786 | 1.14 |
ENSRNOT00000051309
|
Car6
|
carbonic anhydrase 6 |
| chr5_+_57738321 | 1.00 |
ENSRNOT00000063849
|
Ubap1
|
ubiquitin-associated protein 1 |
| chr1_-_219853329 | 0.97 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr4_+_71836677 | 0.92 |
ENSRNOT00000024048
|
Tas2r126
|
taste receptor, type 2, member 126 |
| chr5_+_150032999 | 0.79 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr1_-_222177421 | 0.77 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr1_+_144601410 | 0.53 |
ENSRNOT00000047408
|
Efl1
|
elongation factor like GTPase 1 |
| chr2_+_211050360 | 0.08 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppard
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 5.4 | 21.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 5.3 | 21.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 3.7 | 40.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 2.8 | 17.0 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 2.2 | 15.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 2.0 | 6.1 | GO:0061228 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.9 | 5.7 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 1.8 | 12.7 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 1.7 | 5.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 1.6 | 6.6 | GO:0034696 | response to prostaglandin F(GO:0034696) ketone body biosynthetic process(GO:0046951) |
| 1.0 | 14.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.8 | 13.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.8 | 2.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.5 | 2.5 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.5 | 2.5 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.5 | 4.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 4.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 9.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 8.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.3 | 1.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 5.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 4.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.7 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 2.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.1 | 15.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.7 | 9.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.7 | 21.7 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.6 | 2.5 | GO:0071914 | microspike(GO:0044393) prominosome(GO:0071914) |
| 0.5 | 1.6 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.5 | 11.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 32.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 9.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 4.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 5.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 22.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 17.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 16.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 21.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.1 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 5.7 | 17.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 5.3 | 21.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 3.6 | 21.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 3.4 | 10.2 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 3.2 | 9.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.8 | 12.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.5 | 4.4 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.3 | 6.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.1 | 10.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.9 | 14.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 9.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.4 | 4.8 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.4 | 1.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 2.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 5.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 5.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 15.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 8.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 11.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 6.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 5.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 2.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 32.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.9 | 17.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.4 | 31.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 1.3 | 18.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.0 | 12.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 4.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 24.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 5.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 13.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 5.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 6.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 12.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |