Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pou6f2_Pou4f2
Z-value: 1.09
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
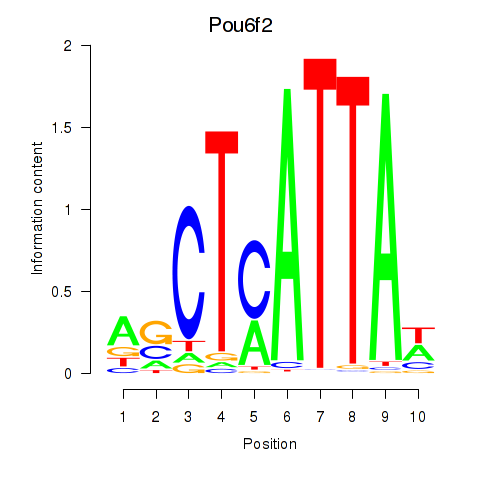
Pou6f2
|
ENSRNOG00000013237 | POU domain, class 6, transcription factor 2 |
|
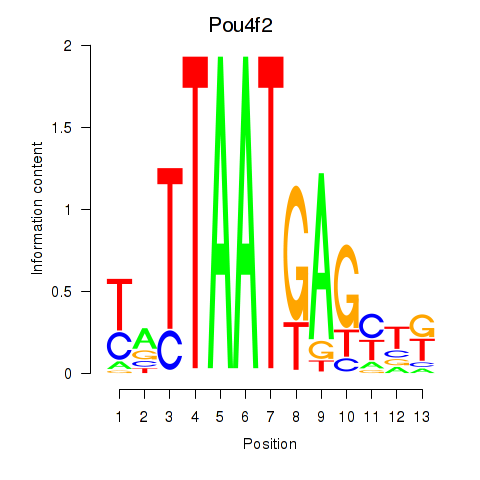
Pou4f2
|
ENSRNOG00000012167 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f2 | rn6_v1_chr19_+_33160180_33160180 | 0.67 | 2.5e-43 | Click! |
| Pou6f2 | rn6_v1_chr17_+_49322205_49322205 | 0.38 | 1.3e-12 | Click! |
Activity profile of Pou6f2_Pou4f2 motif
Sorted Z-values of Pou6f2_Pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_73378057 | 114.81 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chrX_-_142164220 | 60.71 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr19_-_58735173 | 55.14 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr6_+_8284878 | 48.62 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr1_+_234363994 | 42.68 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr16_+_23668595 | 39.35 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr15_+_1054937 | 38.25 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr17_+_9109731 | 37.90 |
ENSRNOT00000016009
|
Cxcl14
|
C-X-C motif chemokine ligand 14 |
| chr6_-_67084234 | 37.82 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr4_+_172942020 | 37.76 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr3_-_66417741 | 33.64 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr13_+_71107465 | 33.56 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr3_+_48106099 | 33.22 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr5_-_68059933 | 32.26 |
ENSRNOT00000088716
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr6_+_73553210 | 31.87 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr1_+_59156251 | 31.66 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr5_-_109651730 | 31.01 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr6_+_83083740 | 29.38 |
ENSRNOT00000007600
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr12_-_2174131 | 27.67 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr5_+_139790395 | 26.75 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr2_+_235596907 | 25.57 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr3_-_81304181 | 24.15 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr8_-_132753145 | 22.73 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chr4_+_108301129 | 22.41 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chrX_-_142248369 | 21.07 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr13_-_76049363 | 18.02 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_-_12526962 | 17.50 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr1_+_157920786 | 16.92 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr2_+_145174876 | 16.56 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr13_+_90723092 | 16.51 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr19_+_6046665 | 16.42 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr17_-_84247038 | 15.81 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chrX_+_84064427 | 15.27 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr13_-_60567882 | 14.95 |
ENSRNOT00000004701
|
Trove2
|
TROVE domain family, member 2 |
| chr7_-_134722215 | 14.89 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr2_+_116970344 | 14.57 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chrX_-_6620722 | 13.77 |
ENSRNOT00000066674
|
Maoa
|
monoamine oxidase A |
| chr15_-_51485692 | 13.58 |
ENSRNOT00000023876
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr15_+_67555835 | 13.26 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chrX_+_86126157 | 13.15 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr8_+_13796021 | 13.06 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr2_+_207930796 | 12.96 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr2_+_113984646 | 12.87 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr13_-_74923402 | 12.84 |
ENSRNOT00000033324
|
Rasal2
|
RAS protein activator like 2 |
| chr5_+_18901039 | 12.43 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr13_+_113373578 | 11.18 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr8_-_45137893 | 11.07 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr10_+_55927223 | 10.69 |
ENSRNOT00000011523
|
Kcnab3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr2_-_5577369 | 10.15 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_97349454 | 9.74 |
ENSRNOT00000089524
|
Dcdc5
|
doublecortin domain containing 5 |
| chr1_-_87825333 | 9.62 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr19_-_24614019 | 9.30 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr2_+_239415046 | 8.73 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr8_-_39551700 | 8.61 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_+_190666149 | 8.32 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr5_-_7941822 | 8.24 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_40879747 | 8.03 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
| chr1_-_72311856 | 7.26 |
ENSRNOT00000021286
|
Epn1
|
Epsin 1 |
| chr1_-_279277339 | 6.63 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr3_+_100770975 | 6.46 |
ENSRNOT00000089233
|
Bdnf
|
brain-derived neurotrophic factor |
| chr18_+_30880020 | 6.01 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr2_-_32518643 | 5.72 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_187786700 | 5.27 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr17_+_26808330 | 5.05 |
ENSRNOT00000022108
|
Bloc1s5
|
biogenesis of lysosomal organelles complex 1 subunit 5 |
| chr5_+_152680407 | 4.86 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr6_+_146784915 | 4.74 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr1_+_217345154 | 4.73 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_8038665 | 4.66 |
ENSRNOT00000046539
|
Aig1
|
androgen-induced 1 |
| chr3_+_104816987 | 4.35 |
ENSRNOT00000042103
ENSRNOT00000044625 |
Fmn1
|
formin 1 |
| chr15_+_4064706 | 4.04 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr5_-_144345531 | 4.04 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr8_-_43854765 | 3.83 |
ENSRNOT00000081506
|
Olr1335
|
olfactory receptor 1335 |
| chr9_-_70787913 | 3.80 |
ENSRNOT00000072007
ENSRNOT00000017901 |
Klf7
Klf7
|
Kruppel like factor 7 Kruppel like factor 7 |
| chr3_-_37803112 | 3.43 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr14_-_18704059 | 3.30 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr15_-_27819376 | 3.06 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr20_+_45458558 | 2.93 |
ENSRNOT00000000713
|
Cdk19
|
cyclin-dependent kinase 19 |
| chr1_-_44474674 | 2.69 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr10_+_53740841 | 2.40 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr3_+_75265525 | 2.35 |
ENSRNOT00000013319
|
Olr554
|
olfactory receptor 554 |
| chr11_-_62067655 | 2.33 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_10577068 | 2.13 |
ENSRNOT00000001282
|
Rnf6
|
ring finger protein 6 |
| chr4_+_68656928 | 1.87 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr11_-_86303453 | 1.80 |
ENSRNOT00000071453
|
LOC498122
|
similar to CG15908-PA |
| chr13_+_80464348 | 1.78 |
ENSRNOT00000076324
|
Vamp4
|
vesicle-associated membrane protein 4 |
| chr12_-_17186679 | 1.46 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr18_-_26656879 | 1.46 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr9_+_71915421 | 1.33 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr4_+_181315444 | 1.32 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr7_+_15628665 | 0.44 |
ENSRNOT00000071081
|
Olr1091
|
olfactory receptor 1091 |
| chr15_-_27485639 | 0.16 |
ENSRNOT00000041030
|
Olr1631
|
olfactory receptor 1631 |
| chr1_-_14117021 | 0.02 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou6f2_Pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 81.8 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 8.0 | 31.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 6.7 | 33.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 6.3 | 37.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 5.5 | 16.5 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 5.3 | 15.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 4.7 | 37.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 3.9 | 42.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 3.1 | 114.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 2.8 | 22.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 2.7 | 21.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 2.6 | 33.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 2.3 | 25.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 2.2 | 24.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 2.0 | 13.8 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 1.7 | 33.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.7 | 5.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.7 | 14.9 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 1.6 | 39.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.6 | 11.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 1.2 | 4.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.2 | 13.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 1.1 | 13.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 1.1 | 3.3 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.1 | 4.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.0 | 31.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.9 | 9.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.9 | 12.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.8 | 6.5 | GO:0061193 | taste bud development(GO:0061193) |
| 0.8 | 8.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 31.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.8 | 10.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.7 | 26.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 2.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.4 | 4.7 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.3 | 48.6 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.2 | 16.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.2 | 4.0 | GO:0060294 | inner dynein arm assembly(GO:0036159) cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 33.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.2 | 2.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 2.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 9.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.2 | 8.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.8 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.1 | 10.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 14.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 2.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 8.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 4.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 6.6 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 | 16.6 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.1 | 1.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 10.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 20.9 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 13.6 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 2.9 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 5.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 5.3 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 12.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.4 | 148.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 10.6 | 31.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 8.0 | 24.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.8 | 7.3 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 1.1 | 16.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.0 | 69.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.9 | 12.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.8 | 16.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.8 | 33.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.6 | 13.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 53.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.5 | 26.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 5.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 6.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 2.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 33.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 5.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 15.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 13.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 8.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 22.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 13.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 7.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 52.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 9.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 49.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 13.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 18.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 9.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 22.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 14.9 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 12.4 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 3.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 16.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 5.0 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 42.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 5.0 | 114.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 4.1 | 16.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 3.5 | 31.9 | GO:0043495 | protein anchor(GO:0043495) |
| 3.4 | 24.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 2.9 | 32.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 2.6 | 13.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 2.1 | 14.9 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.8 | 81.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.5 | 39.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.3 | 10.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.3 | 13.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.2 | 33.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.9 | 2.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.9 | 15.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 3.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.8 | 33.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.8 | 6.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.8 | 11.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.7 | 33.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.7 | 7.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.7 | 25.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.6 | 8.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 8.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 4.7 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 13.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 8.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 22.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 1.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 37.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 27.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 10.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 12.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 2.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 8.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 40.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 6.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 5.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 64.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 13.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 8.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 4.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 16.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 6.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 4.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 114.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.8 | 24.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.5 | 25.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.5 | 4.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 107.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 6.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 12.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 7.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 6.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 4.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 8.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 6.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 48.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.2 | 33.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.0 | 13.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.8 | 52.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.6 | 11.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 16.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.6 | 16.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 23.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 25.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 6.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 7.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 13.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |