Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
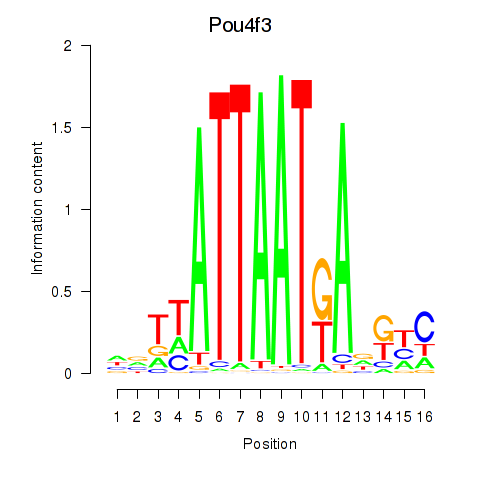
Results for Pou4f3
Z-value: 0.35
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSRNOG00000018842 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f3 | rn6_v1_chr18_+_36713869_36713869 | 0.08 | 1.6e-01 | Click! |
Activity profile of Pou4f3 motif
Sorted Z-values of Pou4f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_142164220 | 8.07 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr3_+_171213936 | 8.05 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr17_-_43543172 | 7.43 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr3_+_159368273 | 7.21 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_253185533 | 6.43 |
ENSRNOT00000067822
|
Pank1
|
pantothenate kinase 1 |
| chr14_-_19072677 | 6.41 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr17_+_9109731 | 5.94 |
ENSRNOT00000016009
|
Cxcl14
|
C-X-C motif chemokine ligand 14 |
| chr18_+_17043903 | 5.42 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr20_+_34258791 | 5.37 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr14_-_84334066 | 5.36 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr1_-_189182306 | 5.34 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr7_-_50278842 | 5.01 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr1_-_189181901 | 5.00 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr9_+_73418607 | 4.99 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr17_-_79085076 | 4.90 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr9_+_73378057 | 4.76 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chrX_+_20520034 | 4.74 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr12_+_41486076 | 4.38 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr12_-_5685448 | 3.60 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr1_-_148119857 | 3.56 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr18_+_14757679 | 3.47 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_+_14835822 | 3.30 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr10_-_89088993 | 3.28 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chrX_-_40086870 | 2.73 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr10_+_103206014 | 2.66 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr20_+_10265806 | 2.42 |
ENSRNOT00000001564
ENSRNOT00000086272 |
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr18_+_30840868 | 2.24 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr12_-_35979193 | 2.18 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_-_27287605 | 2.12 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr19_-_24614019 | 2.11 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr5_+_165724027 | 2.09 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr6_+_127327959 | 2.04 |
ENSRNOT00000012296
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chrX_+_17016778 | 1.87 |
ENSRNOT00000003989
|
Bmp15
|
bone morphogenetic protein 15 |
| chr5_+_145311375 | 1.80 |
ENSRNOT00000019224
|
Smim12
|
small integral membrane protein 12 |
| chr5_-_12199283 | 1.72 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr2_+_239415046 | 1.60 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr1_+_79631668 | 1.48 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr7_+_117456039 | 1.46 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr2_-_105089659 | 1.43 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr3_-_103460529 | 1.38 |
ENSRNOT00000047274
|
LOC100909573
|
olfactory receptor 4F6-like |
| chrX_-_76925195 | 1.36 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr12_-_39697465 | 1.32 |
ENSRNOT00000078683
|
Vps29
|
VPS29 retromer complex component |
| chr7_-_69982592 | 1.27 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr18_+_30017918 | 1.26 |
ENSRNOT00000079794
|
Pcdha4
|
protocadherin alpha 4 |
| chr18_+_30895831 | 1.20 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr1_+_65292386 | 1.20 |
ENSRNOT00000029893
|
AABR07002044.1
|
|
| chrX_-_64715823 | 1.20 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr10_+_29606748 | 1.18 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr3_-_80933283 | 1.18 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chrX_+_65040934 | 1.15 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr16_-_75107931 | 1.15 |
ENSRNOT00000058066
|
Defb15
|
defensin beta 15 |
| chr12_+_10577068 | 1.14 |
ENSRNOT00000001282
|
Rnf6
|
ring finger protein 6 |
| chr4_-_18396035 | 1.13 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr4_-_72020642 | 1.12 |
ENSRNOT00000075946
|
Fam115e
|
family with sequence similarity 115, member E |
| chrX_+_14019961 | 1.09 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr19_+_6046665 | 1.07 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr16_+_75122996 | 0.93 |
ENSRNOT00000018375
|
Defb15
|
defensin beta 15 |
| chr8_-_18910837 | 0.89 |
ENSRNOT00000044295
|
Olr1126
|
olfactory receptor 1126 |
| chr7_+_71157664 | 0.88 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_172348583 | 0.86 |
ENSRNOT00000041144
|
Olr250
|
olfactory receptor 250 |
| chr16_-_20562399 | 0.83 |
ENSRNOT00000031692
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr10_-_60309306 | 0.82 |
ENSRNOT00000083439
|
Olr1486
|
olfactory receptor 1486 |
| chr16_+_84465656 | 0.79 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr6_-_61405195 | 0.76 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr1_-_167875501 | 0.71 |
ENSRNOT00000078322
|
Olr62
|
olfactory receptor 62 |
| chr1_-_168555382 | 0.67 |
ENSRNOT00000091916
|
AC129004.1
|
|
| chr3_+_55910177 | 0.64 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr13_-_102857551 | 0.61 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr3_-_78271875 | 0.61 |
ENSRNOT00000008475
|
Olr701
|
olfactory receptor 701 |
| chr10_-_34301197 | 0.60 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr18_+_56544652 | 0.60 |
ENSRNOT00000024171
|
Pde6a
|
phosphodiesterase 6A |
| chr3_-_74701712 | 0.60 |
ENSRNOT00000041127
|
Olr541
|
olfactory receptor 541 |
| chr17_-_4454701 | 0.51 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr18_-_43945273 | 0.48 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr3_-_103217528 | 0.45 |
ENSRNOT00000074456
|
Olr784
|
olfactory receptor 784 |
| chr1_+_213374895 | 0.39 |
ENSRNOT00000041052
|
Olr311
|
olfactory receptor 311 |
| chr1_-_167884690 | 0.38 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr1_-_168587241 | 0.35 |
ENSRNOT00000021254
|
Olr104
|
olfactory receptor 104 |
| chr8_-_126390801 | 0.33 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr12_-_46493203 | 0.24 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr1_-_190370499 | 0.23 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr14_-_194072 | 0.16 |
ENSRNOT00000073973
|
Vmn2r112
|
vomeronasal 2, receptor 112 |
| chr16_+_48513432 | 0.13 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr18_+_73157973 | 0.10 |
ENSRNOT00000031993
|
Skor2
|
SKI family transcriptional corepressor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.0 | 8.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.2 | 8.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.0 | 5.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 3.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.7 | 2.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.7 | 7.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 1.4 | GO:0035127 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.4 | 1.9 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.4 | 1.5 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.4 | 5.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 6.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 5.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 1.1 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.3 | 9.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 1.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 2.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 3.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 4.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 5.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.6 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 2.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 3.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 6.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 2.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.6 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 2.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 5.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.5 | 2.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 1.4 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 1.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 5.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 7.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 11.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 8.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 6.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 1.0 | 5.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 7.4 | GO:0015562 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 9.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 8.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 7.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 3.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 8.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 5.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 11.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 7.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 8.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 14.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.5 | 6.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 5.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |