Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pou4f1_Pou6f1
Z-value: 0.94
Transcription factors associated with Pou4f1_Pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
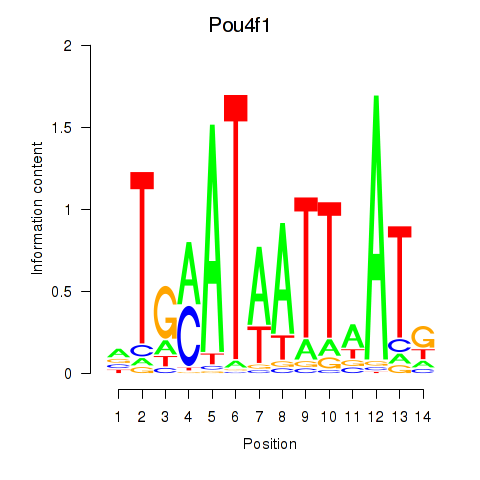
Pou4f1
|
ENSRNOG00000060662 | POU class 4 homeobox 1 |
|
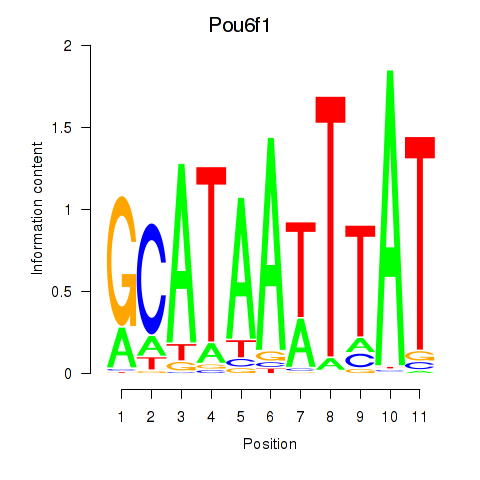
Pou6f1
|
ENSRNOG00000004595 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f1 | rn6_v1_chr15_-_88622413_88622413 | 0.37 | 1.3e-11 | Click! |
| Pou6f1 | rn6_v1_chr7_-_142210738_142210738 | 0.25 | 6.0e-06 | Click! |
Activity profile of Pou4f1_Pou6f1 motif
Sorted Z-values of Pou4f1_Pou6f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_133400485 | 47.56 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr3_-_26056818 | 37.22 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr14_+_60764409 | 33.95 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr1_-_93949187 | 33.12 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr6_+_64789940 | 32.40 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr13_+_57243877 | 31.18 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chrX_-_142164220 | 29.54 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_+_57947716 | 29.28 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr6_-_23291568 | 28.95 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr2_-_196402106 | 28.36 |
ENSRNOT00000028664
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr17_-_43675934 | 25.52 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr14_+_39964588 | 25.36 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr10_-_93675991 | 25.33 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr7_-_50278842 | 24.06 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr16_-_81880502 | 23.58 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr1_-_280015358 | 23.03 |
ENSRNOT00000024548
|
Hspa12a
|
heat shock protein family A (Hsp70) member 12A |
| chr10_+_86303727 | 21.62 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr16_+_39827749 | 21.59 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr11_+_66316606 | 20.78 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr10_-_27366665 | 20.60 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr1_-_25839198 | 20.33 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr2_-_35104963 | 20.10 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr3_+_53563194 | 20.04 |
ENSRNOT00000048300
|
Xirp2
|
xin actin-binding repeat containing 2 |
| chrX_+_53053609 | 19.65 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr15_-_76789298 | 19.61 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr2_-_104461863 | 19.20 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr8_-_80631873 | 18.67 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr7_+_44009069 | 18.25 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr7_-_124929025 | 17.79 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr10_-_47725172 | 17.71 |
ENSRNOT00000003228
|
Rnf112
|
ring finger protein 112 |
| chr16_-_18643309 | 17.45 |
ENSRNOT00000031680
|
Dydc2
|
DPY30 domain containing 2 |
| chr3_+_177188044 | 17.30 |
ENSRNOT00000022073
|
Tcea2
|
transcription elongation factor A2 |
| chr4_+_22859622 | 17.18 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr15_+_1054937 | 17.05 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr9_+_73334618 | 16.98 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_57947183 | 16.86 |
ENSRNOT00000060642
|
Fam219a
|
family with sequence similarity 219, member A |
| chr9_-_66845403 | 16.50 |
ENSRNOT00000041546
|
Ica1l
|
islet cell autoantigen 1-like |
| chr2_+_127625683 | 15.95 |
ENSRNOT00000015474
|
Hspa4l
|
heat shock protein 4-like |
| chr3_+_94848823 | 15.81 |
ENSRNOT00000041362
|
Ccdc73
|
coiled-coil domain containing 73 |
| chr2_-_247446882 | 14.65 |
ENSRNOT00000021963
|
Bmpr1b
|
bone morphogenetic protein receptor type 1B |
| chr2_+_242882306 | 14.59 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr12_-_35979193 | 14.52 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr16_+_7292096 | 14.49 |
ENSRNOT00000025606
|
Tnnc1
|
troponin C1, slow skeletal and cardiac type |
| chr19_+_20607507 | 14.43 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr20_-_25826658 | 14.02 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr11_+_76147205 | 13.95 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr11_+_82194657 | 13.75 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr6_-_23542928 | 13.65 |
ENSRNOT00000083858
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr2_+_69415057 | 13.55 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr14_+_114152472 | 13.38 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr15_-_104168564 | 13.30 |
ENSRNOT00000093385
ENSRNOT00000038596 |
Dzip1
|
DAZ interacting zinc finger protein 1 |
| chr11_+_36851038 | 13.08 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr3_-_66885085 | 13.02 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr2_-_19808937 | 12.81 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr1_-_173764246 | 12.29 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr1_+_137799185 | 11.97 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr6_+_7900972 | 11.68 |
ENSRNOT00000006953
ENSRNOT00000068030 |
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr14_-_55081551 | 11.64 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr1_-_246010594 | 11.62 |
ENSRNOT00000023762
|
Rfx3
|
regulatory factor X3 |
| chrX_-_56765893 | 11.54 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr6_+_27328406 | 11.35 |
ENSRNOT00000091159
ENSRNOT00000044278 |
Otof
|
otoferlin |
| chr3_-_150064438 | 11.33 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chr18_+_30581530 | 11.28 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr15_+_87886783 | 11.21 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr8_-_8524643 | 11.06 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr14_-_43143973 | 10.97 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr14_-_62595854 | 10.87 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr9_-_105376935 | 10.76 |
ENSRNOT00000043386
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr16_-_54628458 | 10.75 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr5_+_103479767 | 10.56 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr9_+_24095751 | 10.40 |
ENSRNOT00000018177
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr13_+_42220251 | 10.31 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr10_-_21265026 | 10.30 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr7_+_141346398 | 10.27 |
ENSRNOT00000077175
|
Asic1
|
acid sensing ion channel subunit 1 |
| chrX_+_159158194 | 10.15 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr3_-_158328881 | 10.11 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr2_+_260039651 | 10.10 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr14_-_41786084 | 9.97 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr1_+_211582077 | 9.97 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr18_+_54108493 | 9.82 |
ENSRNOT00000029239
|
RGD1312005
|
similar to DD1 |
| chr12_+_38368693 | 9.80 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr3_+_161121697 | 9.77 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr15_-_90175802 | 9.72 |
ENSRNOT00000013342
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr14_-_28967980 | 9.67 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr2_-_84531192 | 9.64 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr18_+_30562178 | 9.53 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr8_-_96547568 | 9.35 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr8_+_122076759 | 9.29 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr2_+_92574038 | 9.15 |
ENSRNOT00000089422
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr3_+_148510779 | 9.11 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chr3_-_147188929 | 9.01 |
ENSRNOT00000055455
|
Rad21l1
|
RAD21 cohesin complex component like 1 |
| chr5_+_27326762 | 8.99 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr1_-_16203838 | 8.98 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr10_+_53778662 | 8.97 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr17_+_60287203 | 8.96 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chr14_+_7171613 | 8.93 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr8_+_100260049 | 8.76 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr7_+_11582984 | 8.63 |
ENSRNOT00000026893
|
Gng7
|
G protein subunit gamma 7 |
| chr9_-_114282799 | 8.60 |
ENSRNOT00000090539
|
LOC102555328
|
uncharacterized LOC102555328 |
| chr2_-_137153551 | 8.54 |
ENSRNOT00000083443
|
AABR07010476.1
|
|
| chr11_+_32440237 | 8.44 |
ENSRNOT00000040844
|
Kcne2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr15_-_80713153 | 8.44 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr4_-_135069970 | 8.36 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr12_+_41486076 | 8.07 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr7_+_123043503 | 7.98 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr2_-_219262901 | 7.96 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr6_-_95934296 | 7.93 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr5_-_156689415 | 7.81 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr7_+_15785410 | 7.77 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr16_+_29674793 | 7.76 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr2_-_59181863 | 7.71 |
ENSRNOT00000079636
|
Spef2
|
sperm flagellar 2 |
| chr3_-_160922341 | 7.63 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr9_-_104870382 | 7.56 |
ENSRNOT00000084398
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr6_-_25507073 | 7.53 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr4_-_41212072 | 7.46 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr14_-_108372068 | 7.46 |
ENSRNOT00000088116
ENSRNOT00000091143 ENSRNOT00000085886 |
RGD1305110
|
similar to KIAA1841 protein |
| chr1_-_145931583 | 7.45 |
ENSRNOT00000016433
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr9_+_77320726 | 7.41 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr19_-_41349681 | 7.25 |
ENSRNOT00000080694
ENSRNOT00000059147 |
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr6_+_101603319 | 7.25 |
ENSRNOT00000030470
|
Gphn
|
gephyrin |
| chr14_+_71533063 | 7.24 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr1_-_227392054 | 7.12 |
ENSRNOT00000054806
|
Ms4a13-ps1
|
membrane-spanning 4-domains, subfamily A, member 13, pseudogene 1 |
| chr14_+_84231639 | 7.06 |
ENSRNOT00000066362
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr9_+_47281961 | 7.02 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr19_+_25143428 | 6.82 |
ENSRNOT00000007535
|
Palm3
|
paralemmin 3 |
| chr8_+_55504359 | 6.79 |
ENSRNOT00000059086
|
Btg4
|
BTG anti-proliferation factor 4 |
| chr2_-_186232292 | 6.73 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr1_+_101603222 | 6.71 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr1_+_172752416 | 6.63 |
ENSRNOT00000087448
|
Olr268
|
olfactory receptor 268 |
| chr3_+_154395187 | 6.59 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr10_+_46722109 | 6.55 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chr1_-_227486976 | 6.51 |
ENSRNOT00000035812
|
Ms4a14
|
membrane spanning 4-domains A14 |
| chr7_-_68549763 | 6.39 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr19_-_22194740 | 6.35 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr14_-_92077088 | 6.32 |
ENSRNOT00000085175
|
Grb10
|
growth factor receptor bound protein 10 |
| chr10_-_89187474 | 6.31 |
ENSRNOT00000064931
|
Usmg5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr2_+_113984646 | 6.27 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr7_-_120380200 | 6.24 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr8_+_107882219 | 6.22 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr10_-_57671080 | 6.22 |
ENSRNOT00000082511
|
LOC691995
|
hypothetical protein LOC691995 |
| chr1_+_162890475 | 6.14 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr10_-_36322356 | 6.06 |
ENSRNOT00000000248
|
Zfp354c
|
zinc finger protein 354C |
| chr3_+_112428395 | 6.06 |
ENSRNOT00000079109
ENSRNOT00000048141 |
Stard9
|
StAR-related lipid transfer domain containing 9 |
| chr15_-_2731462 | 6.03 |
ENSRNOT00000018023
|
Samd8
|
sterile alpha motif domain containing 8 |
| chr4_-_163095614 | 5.97 |
ENSRNOT00000088759
|
RGD1564770
|
similar to CD69 antigen (p60, early T-cell activation antigen) |
| chr10_+_53818818 | 5.92 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr9_-_32868371 | 5.85 |
ENSRNOT00000038369
|
LOC689725
|
similar to chromosome 9 open reading frame 79 |
| chr7_-_106695570 | 5.78 |
ENSRNOT00000083517
|
Hhla1
|
HERV-H LTR-associating 1 |
| chr5_-_12563429 | 5.74 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr8_+_71216178 | 5.70 |
ENSRNOT00000021372
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr7_+_25808419 | 5.69 |
ENSRNOT00000091571
|
Rfx4
|
regulatory factor X4 |
| chr18_+_29987206 | 5.64 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_205030567 | 5.45 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
| chr17_+_77176716 | 5.44 |
ENSRNOT00000083342
ENSRNOT00000024162 |
Optn
|
optineurin |
| chr1_+_5448958 | 5.41 |
ENSRNOT00000061930
|
Epm2a
|
epilepsy, progressive myoclonus type 2A |
| chr16_-_20641908 | 5.26 |
ENSRNOT00000026846
|
Ell
|
elongation factor for RNA polymerase II |
| chr17_-_79085076 | 5.24 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr18_-_38374 | 5.23 |
ENSRNOT00000036914
|
Vom2r75
|
vomeronasal 2 receptor, 75 |
| chr1_-_100669684 | 5.22 |
ENSRNOT00000091760
|
Myh14
|
myosin heavy chain 14 |
| chr5_-_6186329 | 5.19 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr15_+_3938075 | 5.17 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr18_+_30424814 | 5.14 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr15_-_35153804 | 5.12 |
ENSRNOT00000050660
|
Gzmn
|
granzyme N |
| chr2_-_88763733 | 5.07 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr10_+_31508512 | 5.00 |
ENSRNOT00000031580
|
Fam71b
|
family with sequence similarity 71, member B |
| chr13_-_74923402 | 4.95 |
ENSRNOT00000033324
|
Rasal2
|
RAS protein activator like 2 |
| chr14_-_1505085 | 4.92 |
ENSRNOT00000090361
|
LOC102554799
|
uncharacterized LOC102554799 |
| chrX_-_74968405 | 4.89 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr2_-_132301073 | 4.88 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr9_+_53013413 | 4.79 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr3_+_100786862 | 4.75 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr20_-_34684418 | 4.74 |
ENSRNOT00000087220
|
Cep85l
|
centrosomal protein 85-like |
| chr7_-_122926336 | 4.72 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr18_+_30509393 | 4.65 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr9_+_101268905 | 4.65 |
ENSRNOT00000023270
|
Olr178
|
olfactory receptor 178 |
| chr2_+_92549479 | 4.59 |
ENSRNOT00000082912
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chrX_+_63542191 | 4.56 |
ENSRNOT00000073955
|
Apoo
|
apolipoprotein O |
| chr13_-_47916185 | 4.54 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr17_+_49417067 | 4.49 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr9_-_60672246 | 4.46 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_-_49844547 | 4.42 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr19_+_43163129 | 4.36 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chrX_-_122062799 | 4.29 |
ENSRNOT00000086852
|
Gm4907
|
predicted gene 4907 |
| chr8_+_49077053 | 4.28 |
ENSRNOT00000087922
|
Ift46
|
intraflagellar transport 46 |
| chr5_-_153840178 | 4.27 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr9_-_88816898 | 4.27 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr8_+_99680406 | 4.21 |
ENSRNOT00000042834
|
LOC102551265
|
phospholipid scramblase 1-like |
| chr20_+_17750744 | 4.15 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr20_-_776472 | 4.10 |
ENSRNOT00000049250
|
Olr1690
|
olfactory receptor 1690 |
| chr9_+_24066303 | 3.99 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr3_-_160561741 | 3.93 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr1_+_1702696 | 3.92 |
ENSRNOT00000019181
|
Lrp11
|
LDL receptor related protein 11 |
| chr1_-_66321642 | 3.91 |
ENSRNOT00000074234
|
AABR07002093.1
|
|
| chr10_-_36323055 | 3.88 |
ENSRNOT00000081026
|
Zfp354c
|
zinc finger protein 354C |
| chr3_+_102456938 | 3.85 |
ENSRNOT00000051827
|
Olr748
|
olfactory receptor 748 |
| chr15_+_4850122 | 3.84 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr6_+_102215603 | 3.80 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr16_+_90325304 | 3.78 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr1_-_48797631 | 3.76 |
ENSRNOT00000082958
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_-_47477524 | 3.72 |
ENSRNOT00000067916
|
Rsph3
|
radial spoke 3 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f1_Pou6f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 6.5 | 19.6 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 4.8 | 14.5 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 4.8 | 19.2 | GO:0070093 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of cortisol secretion(GO:0051464) negative regulation of glucagon secretion(GO:0070093) |
| 4.2 | 29.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 3.9 | 11.6 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 3.7 | 33.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 3.6 | 32.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 3.3 | 13.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 3.1 | 21.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 3.1 | 12.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 2.8 | 8.4 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 2.8 | 14.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.8 | 13.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 2.7 | 10.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 2.6 | 7.9 | GO:2000729 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 2.5 | 7.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 2.5 | 7.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.4 | 7.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 2.3 | 11.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 2.2 | 13.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 2.2 | 28.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 2.1 | 38.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 2.1 | 14.7 | GO:0022605 | oogenesis stage(GO:0022605) |
| 2.0 | 12.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 2.0 | 9.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 1.9 | 5.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.9 | 18.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.8 | 7.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.8 | 14.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.7 | 17.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 1.7 | 5.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.7 | 24.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.7 | 18.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.7 | 11.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.6 | 8.2 | GO:0098970 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 1.6 | 9.7 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.6 | 8.0 | GO:0061743 | motor learning(GO:0061743) |
| 1.6 | 7.8 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) maintenance of protein location in mitochondrion(GO:0072656) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 1.5 | 20.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.5 | 10.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.4 | 11.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.4 | 4.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 1.3 | 10.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.3 | 6.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.2 | 8.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 1.2 | 14.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.2 | 3.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 1.1 | 5.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.1 | 4.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.1 | 3.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 1.1 | 5.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.1 | 3.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 1.0 | 9.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.0 | 7.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 1.0 | 13.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.0 | 5.9 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 1.0 | 4.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 1.0 | 2.9 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.9 | 7.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.9 | 10.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.9 | 25.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.9 | 2.7 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.9 | 5.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.9 | 11.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.9 | 32.2 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.9 | 47.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.8 | 9.0 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.8 | 2.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.8 | 4.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.8 | 10.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.8 | 3.1 | GO:0070086 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) |
| 0.8 | 6.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.7 | 23.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.7 | 9.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.7 | 5.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 5.4 | GO:0046959 | habituation(GO:0046959) |
| 0.6 | 6.0 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.6 | 7.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.6 | 7.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.6 | 5.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.5 | 17.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.5 | 9.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.5 | 66.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 6.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 3.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.4 | 8.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.4 | 3.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.4 | 9.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 3.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 13.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.4 | 1.2 | GO:0016107 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.4 | 10.2 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.4 | 3.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 20.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.4 | 12.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 2.3 | GO:0008078 | mesodermal cell migration(GO:0008078) establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 13.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.4 | 3.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 3.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.3 | 1.4 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 1.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.3 | 4.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 4.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.3 | 20.0 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.3 | 2.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 8.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 2.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 25.6 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.3 | 4.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 3.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.3 | 1.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 3.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 2.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 3.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 3.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 3.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 7.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 | 1.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 1.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 5.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 2.3 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.2 | 16.0 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.2 | 10.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 1.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 3.3 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.2 | 1.5 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.2 | 3.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 8.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.7 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 4.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 3.6 | GO:0071450 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 | 5.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 16.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 4.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 4.9 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.1 | 0.3 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.1 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 20.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 4.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 3.9 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 8.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 2.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 2.8 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 3.5 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 2.3 | GO:0060443 | mammary gland morphogenesis(GO:0060443) |
| 0.0 | 0.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 2.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 1.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 3.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.2 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 4.9 | 29.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 4.0 | 24.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 3.6 | 14.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.9 | 14.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 2.3 | 18.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.9 | 11.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.9 | 17.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.9 | 7.5 | GO:0071914 | prominosome(GO:0071914) |
| 1.3 | 9.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.3 | 5.3 | GO:0035363 | histone locus body(GO:0035363) |
| 1.3 | 5.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.3 | 9.0 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.2 | 32.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.2 | 3.6 | GO:1990923 | PET complex(GO:1990923) |
| 1.1 | 11.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.1 | 9.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.1 | 12.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.1 | 19.2 | GO:0043196 | varicosity(GO:0043196) |
| 1.1 | 3.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 1.0 | 9.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.0 | 4.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.0 | 17.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.9 | 20.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.9 | 7.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.8 | 13.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 6.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 14.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.7 | 25.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.7 | 10.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.7 | 7.7 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 10.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 3.5 | GO:0005940 | septin ring(GO:0005940) |
| 0.7 | 4.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 3.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.6 | 8.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.6 | 29.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 10.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.5 | 58.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.5 | 5.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 3.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 13.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.5 | 8.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 3.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 43.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 3.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.4 | 11.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 5.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.4 | 3.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 8.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 3.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 4.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 9.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.3 | 2.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 12.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 4.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 11.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 6.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 25.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 4.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 23.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 6.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 5.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 15.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 4.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 16.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 15.5 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 0.5 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.1 | 2.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 13.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 6.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 21.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 6.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 5.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 9.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 8.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 8.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 5.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 5.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 17.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 21.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 17.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 16.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 3.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.5 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 5.9 | 53.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 4.8 | 24.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.8 | 19.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 4.1 | 33.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 4.1 | 20.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 3.5 | 10.4 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 3.4 | 10.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 3.3 | 13.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.9 | 11.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 2.7 | 11.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.5 | 7.5 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 2.4 | 14.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 2.1 | 18.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.1 | 8.2 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 2.0 | 45.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.9 | 5.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.8 | 14.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 1.8 | 5.4 | GO:2001070 | starch binding(GO:2001070) |
| 1.8 | 7.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.5 | 25.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.4 | 4.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 1.3 | 5.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.3 | 6.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.2 | 6.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.2 | 6.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.1 | 5.4 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 1.1 | 31.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 1.0 | 7.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.0 | 11.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 1.0 | 7.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.9 | 8.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.9 | 43.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.9 | 11.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.9 | 12.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.8 | 10.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 21.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.8 | 6.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 7.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.7 | 17.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.6 | 12.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.6 | 3.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 3.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 9.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 3.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.6 | 8.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 15.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 4.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.5 | 13.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.5 | 11.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.5 | 1.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.4 | 1.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.4 | 4.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 2.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 19.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 3.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 2.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 1.4 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 1.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 3.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 9.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 1.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 4.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 7.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 4.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 2.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 2.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 25.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 4.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 5.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 21.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 6.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 23.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.2 | 5.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 12.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 13.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 10.9 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 5.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 2.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 4.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 36.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 16.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 6.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 6.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 13.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 3.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 12.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 26.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 47.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.6 | 22.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.6 | 26.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 20.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.4 | 32.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 17.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 9.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 6.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 14.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 9.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 7.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 11.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 3.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 6.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 5.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 5.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 31.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 31.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 3.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 1.7 | 24.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.6 | 47.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 1.2 | 20.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.1 | 18.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 1.0 | 40.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.9 | 32.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.9 | 24.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.9 | 11.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.8 | 9.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.6 | 10.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.5 | 12.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.5 | 6.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 14.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 13.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 3.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 13.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 6.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 2.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 3.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 13.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 5.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 16.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 5.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 7.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 9.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 2.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 11.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 11.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 10.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 9.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |