Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
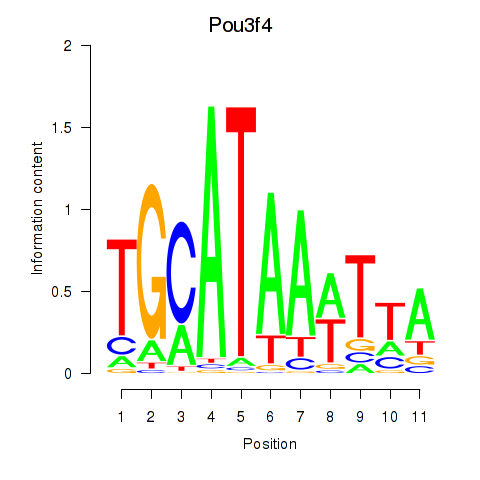
Results for Pou3f4
Z-value: 0.76
Transcription factors associated with Pou3f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f4
|
ENSRNOG00000002784 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f4 | rn6_v1_chrX_+_82143789_82143789 | 0.63 | 8.6e-37 | Click! |
Activity profile of Pou3f4 motif
Sorted Z-values of Pou3f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_156552328 | 27.95 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr15_-_71779033 | 26.86 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr11_+_20474483 | 26.52 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr16_-_81880502 | 23.85 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr1_+_151439409 | 23.22 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr7_+_133400485 | 23.05 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr12_-_29743705 | 21.49 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr11_+_42259761 | 21.44 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr5_+_145257714 | 21.39 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr2_-_19808937 | 20.75 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr3_-_26056818 | 20.08 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr7_-_93502571 | 19.67 |
ENSRNOT00000077033
ENSRNOT00000076080 |
Samd12
|
sterile alpha motif domain containing 12 |
| chr4_-_135069970 | 16.83 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr1_-_93949187 | 16.79 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr9_-_45558993 | 16.56 |
ENSRNOT00000087866
ENSRNOT00000017714 |
Chst10
|
carbohydrate sulfotransferase 10 |
| chr10_+_54155876 | 15.19 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr20_-_25826658 | 15.16 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr10_+_74871523 | 13.09 |
ENSRNOT00000076995
ENSRNOT00000076026 ENSRNOT00000088581 ENSRNOT00000076701 |
Sept4
|
septin 4 |
| chr7_-_124929025 | 12.77 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr14_-_91989307 | 12.74 |
ENSRNOT00000057051
|
Ddc
|
dopa decarboxylase |
| chr19_-_11669578 | 12.44 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr7_+_121408829 | 12.21 |
ENSRNOT00000023434
|
Mgat3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr6_+_102215603 | 11.70 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr18_+_30527705 | 11.52 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr6_+_29518416 | 10.92 |
ENSRNOT00000078660
|
AABR07063346.1
|
|
| chr5_-_16799776 | 10.89 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr13_-_47916185 | 10.71 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr5_-_153840178 | 10.46 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr18_+_30398113 | 10.37 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr2_-_104461863 | 10.21 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr5_-_156689415 | 10.10 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr15_-_80713153 | 9.97 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr6_+_7900972 | 9.56 |
ENSRNOT00000006953
ENSRNOT00000068030 |
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr14_-_55081551 | 8.96 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr2_-_28003186 | 8.89 |
ENSRNOT00000048554
|
Hexb
|
hexosaminidase subunit beta |
| chr14_+_114152472 | 8.57 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr3_-_107760550 | 8.44 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr14_+_60764409 | 8.27 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr6_-_135251073 | 8.24 |
ENSRNOT00000088986
|
Mok
|
MOK protein kinase |
| chr4_-_145678066 | 8.20 |
ENSRNOT00000014103
|
Ghrl
|
ghrelin and obestatin prepropeptide |
| chr11_+_82194657 | 8.20 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr8_+_122076759 | 8.18 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr1_-_49844547 | 8.17 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr2_+_22909569 | 8.13 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr5_+_27326762 | 7.44 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr5_+_57947716 | 7.44 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr1_+_217151166 | 7.38 |
ENSRNOT00000071484
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr20_+_25990304 | 7.28 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_8928534 | 7.15 |
ENSRNOT00000088509
ENSRNOT00000065300 |
Miga2
|
mitoguardin 2 |
| chr2_+_12516702 | 7.07 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr1_+_236031988 | 6.98 |
ENSRNOT00000016164
|
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr1_-_67094567 | 6.56 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr18_+_61377051 | 6.46 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr6_+_44230985 | 6.30 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr8_+_82043403 | 6.06 |
ENSRNOT00000091789
|
Myo5a
|
myosin VA |
| chr5_-_57947183 | 5.56 |
ENSRNOT00000060642
|
Fam219a
|
family with sequence similarity 219, member A |
| chr2_-_259382765 | 5.23 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr7_-_117788550 | 4.76 |
ENSRNOT00000021775
|
MGC94207
|
similar to RIKEN cDNA C030006K11 |
| chr19_+_39126589 | 4.71 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chr13_-_61003744 | 4.47 |
ENSRNOT00000005163
|
Rgs13
|
regulator of G-protein signaling 13 |
| chr1_+_65781547 | 4.33 |
ENSRNOT00000026204
|
Zfp329
|
zinc finger protein 329 |
| chr2_+_72006099 | 4.11 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr2_-_149347796 | 4.07 |
ENSRNOT00000091020
|
P2ry14
|
purinergic receptor P2Y14 |
| chr19_+_21541742 | 3.93 |
ENSRNOT00000021095
ENSRNOT00000042077 |
Abcc12
|
ATP binding cassette subfamily C member 12 |
| chr9_-_66019065 | 3.80 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr15_-_55424024 | 3.73 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr10_-_87335823 | 3.72 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr10_-_59888198 | 3.68 |
ENSRNOT00000093482
ENSRNOT00000049311 ENSRNOT00000093230 |
Aspa
|
aspartoacylase |
| chr7_+_9897983 | 3.67 |
ENSRNOT00000042498
|
LOC100910577
|
zinc finger protein 709-like |
| chr16_+_32457521 | 3.67 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr1_-_66321642 | 3.65 |
ENSRNOT00000074234
|
AABR07002093.1
|
|
| chrM_+_5323 | 3.50 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr11_-_62128044 | 3.44 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_78735279 | 3.21 |
ENSRNOT00000011970
|
Malsu1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr8_+_100260049 | 3.05 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr15_+_18539210 | 3.04 |
ENSRNOT00000083212
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr1_-_251387002 | 2.99 |
ENSRNOT00000092063
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr16_-_9658484 | 2.97 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr6_+_36107672 | 2.96 |
ENSRNOT00000093003
|
Rdh14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr10_-_95431312 | 2.73 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr1_+_61733805 | 2.64 |
ENSRNOT00000086029
|
LOC682206
|
similar to Zinc finger protein 208 |
| chr5_+_64412895 | 2.59 |
ENSRNOT00000010561
|
Murc
|
muscle-related coiled-coil protein |
| chr3_+_72540538 | 2.56 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr14_-_11546314 | 2.50 |
ENSRNOT00000090334
|
AABR07014355.1
|
|
| chr2_-_147693082 | 2.38 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr3_-_50120392 | 2.30 |
ENSRNOT00000006175
|
Fign
|
fidgetin, microtubule severing factor |
| chr10_-_87300728 | 2.28 |
ENSRNOT00000036023
ENSRNOT00000081709 |
Krt10
|
keratin 10 |
| chr18_-_38374 | 2.09 |
ENSRNOT00000036914
|
Vom2r75
|
vomeronasal 2 receptor, 75 |
| chr19_-_41029206 | 2.06 |
ENSRNOT00000023383
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr9_-_81879211 | 1.84 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chr5_+_116420690 | 1.83 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr15_+_12827707 | 1.49 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr7_+_12840938 | 1.49 |
ENSRNOT00000039232
|
Polrmt
|
RNA polymerase mitochondrial |
| chrX_+_136466779 | 1.42 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_+_88298266 | 1.41 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr10_+_83476107 | 1.37 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr3_-_77389521 | 1.29 |
ENSRNOT00000043928
|
Olr657
|
olfactory receptor 657 |
| chr6_-_103949275 | 1.27 |
ENSRNOT00000047825
|
AABR07065012.1
|
|
| chr6_+_106052212 | 1.24 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr2_-_219262901 | 1.18 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr3_-_74701712 | 1.05 |
ENSRNOT00000041127
|
Olr541
|
olfactory receptor 541 |
| chr9_-_32868371 | 0.99 |
ENSRNOT00000038369
|
LOC689725
|
similar to chromosome 9 open reading frame 79 |
| chr4_-_165747201 | 0.96 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr7_-_13108630 | 0.94 |
ENSRNOT00000087038
ENSRNOT00000080491 |
Giot1
|
gonadotropin inducible ovarian transcription factor 1 |
| chr2_+_138194136 | 0.88 |
ENSRNOT00000014928
|
RGD1307595
|
similar to RIKEN cDNA 1700018B24 |
| chr3_-_150960030 | 0.86 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr8_+_13305152 | 0.85 |
ENSRNOT00000012940
|
Mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr1_-_230079916 | 0.82 |
ENSRNOT00000091024
|
Olr358
|
olfactory receptor 358 |
| chr7_-_97071968 | 0.81 |
ENSRNOT00000006596
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr9_-_99821954 | 0.76 |
ENSRNOT00000090382
ENSRNOT00000022220 |
Otos
|
otospiralin |
| chr3_-_78886096 | 0.66 |
ENSRNOT00000008864
|
Olr729
|
olfactory receptor 729 |
| chrX_+_157759624 | 0.53 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr20_-_3440769 | 0.52 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr2_-_170300408 | 0.41 |
ENSRNOT00000046681
|
Si
|
sucrase-isomaltase |
| chr7_-_140506925 | 0.41 |
ENSRNOT00000083354
|
Kmt2d
|
lysine methyltransferase 2D |
| chr13_-_90443157 | 0.40 |
ENSRNOT00000006862
|
Nhlh1
|
nescient helix loop helix 1 |
| chrX_+_110670119 | 0.38 |
ENSRNOT00000014811
ENSRNOT00000089579 |
LOC680663
|
hypothetical protein LOC680663 |
| chr9_+_19451630 | 0.37 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr11_-_28979169 | 0.36 |
ENSRNOT00000061601
|
AABR07033580.1
|
|
| chr5_+_63870432 | 0.35 |
ENSRNOT00000007641
|
Stx17
|
syntaxin 17 |
| chr1_+_172728264 | 0.33 |
ENSRNOT00000075297
|
Olr267
|
olfactory receptor 267 |
| chr1_-_88558696 | 0.26 |
ENSRNOT00000038642
|
AABR07002896.1
|
|
| chr3_-_94418711 | 0.23 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr1_-_166988062 | 0.22 |
ENSRNOT00000039564
|
Lrtomt
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr1_-_101773508 | 0.19 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr8_+_19548488 | 0.10 |
ENSRNOT00000008605
|
Olr1148
|
olfactory receptor 1148 |
| chr3_-_72808425 | 0.01 |
ENSRNOT00000048581
|
Olr441
|
olfactory receptor 441 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 4.6 | 18.4 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 3.2 | 12.7 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 2.6 | 13.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 2.5 | 28.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 2.1 | 10.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 2.0 | 10.1 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 1.9 | 15.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.9 | 16.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.6 | 8.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.5 | 6.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.4 | 9.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.3 | 8.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.2 | 8.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.2 | 7.0 | GO:0002001 | renin secretion into blood stream(GO:0002001) |
| 1.1 | 3.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 1.0 | 3.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.0 | 28.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.9 | 3.7 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.8 | 8.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 10.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.7 | 3.7 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.7 | 24.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.7 | 21.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.7 | 12.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.6 | 7.3 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.6 | 7.4 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.5 | 10.9 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.5 | 3.8 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.5 | 8.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 6.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.5 | 19.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.4 | 61.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 7.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 3.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.4 | 16.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.4 | 4.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.4 | 23.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.3 | 1.4 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.3 | 2.4 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.3 | 5.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.3 | 12.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 10.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.2 | 3.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 7.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 3.0 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 3.7 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 0.8 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 7.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 | 1.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 2.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 7.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 15.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 4.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.4 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 2.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 3.4 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 0.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 1.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.4 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 1.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 2.0 | 6.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.7 | 28.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.7 | 20.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.6 | 33.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.3 | 13.1 | GO:0031105 | septin complex(GO:0031105) sperm annulus(GO:0097227) |
| 1.3 | 21.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.2 | 7.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.2 | 8.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.8 | 15.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.8 | 21.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 7.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 10.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 8.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 7.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 39.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 9.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 3.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 12.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 8.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 3.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 2.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.3 | 1.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 8.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 10.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 23.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 3.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 8.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 15.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 3.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 2.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 46.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 21.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 6.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 36.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 10.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 23.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 4.2 | 12.7 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 3.8 | 22.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 2.8 | 28.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 2.4 | 9.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 2.1 | 16.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 2.1 | 8.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 1.8 | 8.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.7 | 15.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.6 | 8.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.2 | 3.7 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 1.2 | 3.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.2 | 3.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.0 | 5.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.0 | 20.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.0 | 10.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.0 | 23.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.9 | 18.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.8 | 3.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.7 | 10.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.6 | 21.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.6 | 7.4 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.5 | 2.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 1.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.4 | 3.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.4 | 8.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 6.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 4.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 5.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 12.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.3 | 10.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 16.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 23.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.2 | 3.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 3.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 7.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 57.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.4 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 11.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 6.3 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.8 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 1.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 4.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 7.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 9.5 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 3.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 8.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 6.5 | GO:0016747 | transferase activity, transferring acyl groups other than amino-acyl groups(GO:0016747) |
| 0.0 | 8.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 10.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 7.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 7.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.7 | 23.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.6 | 21.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 23.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 12.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 3.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 8.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 8.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 23.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.7 | 8.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 8.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 12.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.4 | 12.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.4 | 5.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 3.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 6.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 6.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 10.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 9.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 8.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.5 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |