Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
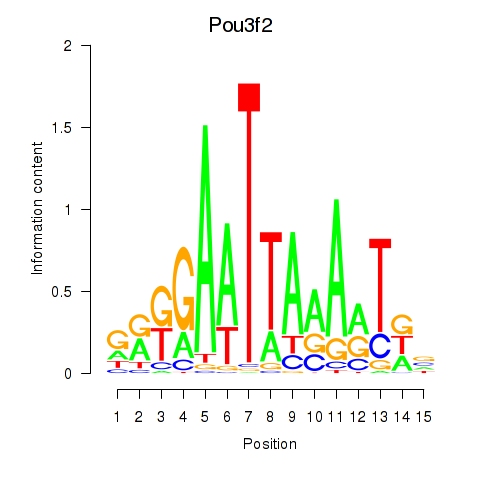
Results for Pou3f2
Z-value: 0.78
Transcription factors associated with Pou3f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f2
|
ENSRNOG00000006908 | POU class 3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f2 | rn6_v1_chr5_-_36683356_36683356 | 0.43 | 4.6e-16 | Click! |
Activity profile of Pou3f2 motif
Sorted Z-values of Pou3f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_106998623 | 42.84 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr9_-_32019205 | 33.44 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_-_159287193 | 30.81 |
ENSRNOT00000080139
|
Kcna6
|
potassium voltage-gated channel subfamily A member 6 |
| chr4_+_21462779 | 29.45 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr17_+_53231343 | 26.79 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr4_+_35279063 | 25.34 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr6_-_23291568 | 25.04 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_130085838 | 24.59 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr4_-_117268178 | 23.31 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr10_+_1920529 | 23.27 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr11_-_11213821 | 19.67 |
ENSRNOT00000093706
|
Robo2
|
roundabout guidance receptor 2 |
| chr4_-_17594598 | 19.57 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr13_+_24823488 | 19.26 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chr7_+_64768742 | 18.90 |
ENSRNOT00000005545
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr17_-_8925503 | 18.49 |
ENSRNOT00000015163
|
Catsper3
|
cation channel, sperm associated 3 |
| chrX_-_56765893 | 18.43 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr11_-_11214141 | 17.85 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chr5_-_172424081 | 16.41 |
ENSRNOT00000019096
|
Plch2
|
phospholipase C, eta 2 |
| chr3_-_111994337 | 15.56 |
ENSRNOT00000030502
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr13_-_47916185 | 15.20 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr17_+_43423111 | 14.94 |
ENSRNOT00000022630
|
Hist1h2ba
|
histone cluster 1 H2B family member a |
| chr5_+_146656049 | 14.23 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr2_+_3662763 | 13.93 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_+_162890475 | 13.54 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr8_+_28454962 | 12.66 |
ENSRNOT00000051573
|
Spata19
|
spermatogenesis associated 19 |
| chr14_-_28536260 | 12.43 |
ENSRNOT00000059942
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr16_-_32753278 | 12.21 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr15_-_71779033 | 12.12 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr10_-_93679974 | 12.09 |
ENSRNOT00000009316
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr17_-_43422846 | 11.98 |
ENSRNOT00000050526
|
Hist1h2aa
|
histone cluster 1 H2A family member a |
| chr15_+_40665041 | 11.34 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr7_-_132143470 | 11.20 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr6_+_126874193 | 11.05 |
ENSRNOT00000011775
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr18_+_51492196 | 10.87 |
ENSRNOT00000020570
|
Gramd3
|
GRAM domain containing 3 |
| chr19_-_56037077 | 10.63 |
ENSRNOT00000021968
|
Spata2L
|
spermatogenesis associated 2-like |
| chr12_+_18931477 | 10.50 |
ENSRNOT00000077087
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr15_-_5667328 | 10.34 |
ENSRNOT00000061522
|
AABR07016976.1
|
|
| chr13_+_93751759 | 10.28 |
ENSRNOT00000065706
|
Wdr64
|
WD repeat domain 64 |
| chr10_+_69737328 | 9.92 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr7_+_34402738 | 9.89 |
ENSRNOT00000030985
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr6_+_78567970 | 9.86 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr2_+_27630285 | 9.54 |
ENSRNOT00000032960
|
Ankrd31
|
ankyrin repeat domain 31 |
| chr10_-_56426012 | 9.47 |
ENSRNOT00000092506
ENSRNOT00000084068 |
LOC497938
|
similar to RIKEN cDNA 4933402P03 |
| chr16_+_52169450 | 9.30 |
ENSRNOT00000019474
|
Triml1
|
tripartite motif family-like 1 |
| chr15_+_58016238 | 8.99 |
ENSRNOT00000075786
|
Kctd4
|
potassium channel tetramerization domain containing 4 |
| chr7_-_27552078 | 8.91 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr5_-_115387377 | 8.63 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr3_+_138770574 | 8.53 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr9_-_110225486 | 8.24 |
ENSRNOT00000045096
|
Efna5
|
ephrin A5 |
| chr5_-_73494030 | 8.09 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr1_+_146976975 | 7.57 |
ENSRNOT00000071895
|
LOC100366231
|
X-linked lymphocyte-regulated 5C-like |
| chr17_+_59714657 | 7.50 |
ENSRNOT00000035675
|
RGD1560860
|
similar to ankyrin repeat domain 26 |
| chrX_-_122062799 | 7.49 |
ENSRNOT00000086852
|
Gm4907
|
predicted gene 4907 |
| chr3_+_54253949 | 7.29 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr5_-_76261198 | 7.28 |
ENSRNOT00000087380
|
AABR07048387.1
|
|
| chr18_-_81682206 | 7.28 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr9_+_10941613 | 7.24 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr10_+_17327275 | 7.10 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr13_-_74520634 | 6.86 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_+_9790422 | 6.80 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr3_+_128155069 | 6.67 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr1_-_67094567 | 6.37 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr2_-_132301073 | 6.25 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr6_-_95796502 | 6.18 |
ENSRNOT00000041409
|
Six6os1
|
Six6 opposite strand transcript 1 |
| chr4_+_8066907 | 6.18 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr9_+_119542328 | 6.16 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr20_+_21316826 | 5.97 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chrX_+_122313470 | 5.80 |
ENSRNOT00000018322
|
RGD1566387
|
similar to hypothetical protein |
| chr1_+_89149998 | 5.51 |
ENSRNOT00000056742
|
LOC688924
|
hypothetical protein LOC688924 |
| chr6_+_91532467 | 5.51 |
ENSRNOT00000082500
ENSRNOT00000064668 |
Klhdc1
|
kelch domain containing 1 |
| chr2_-_216348194 | 5.51 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr10_-_52290657 | 5.44 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr10_-_72472675 | 5.43 |
ENSRNOT00000081649
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr20_+_1197746 | 5.37 |
ENSRNOT00000045794
|
Olr1710
|
olfactory receptor 1710 |
| chr8_-_114010277 | 5.34 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr13_+_67545430 | 5.29 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr5_+_154868534 | 5.09 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr15_+_18941431 | 5.05 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr2_-_173836854 | 4.97 |
ENSRNOT00000074278
|
Wdr49
|
WD repeat domain 49 |
| chr7_-_68523364 | 4.96 |
ENSRNOT00000077670
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr6_+_73553210 | 4.87 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr12_+_10063117 | 4.82 |
ENSRNOT00000044038
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr15_-_83442008 | 4.72 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
| chr10_-_70181708 | 4.39 |
ENSRNOT00000076598
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr2_-_232178533 | 4.25 |
ENSRNOT00000082173
ENSRNOT00000055607 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr10_-_58771908 | 4.25 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chrX_-_61151601 | 4.05 |
ENSRNOT00000004132
|
Mageb4
|
melanoma antigen family B, 4 |
| chr2_+_138489364 | 3.97 |
ENSRNOT00000042248
|
LOC103691563
|
60S ribosomal protein L31 pseudogene |
| chrX_-_915953 | 3.92 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr4_+_8066737 | 3.89 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr7_-_122926336 | 3.88 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr6_-_146470456 | 3.86 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr19_+_43163129 | 3.86 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_+_266844480 | 3.72 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr2_-_142235066 | 3.44 |
ENSRNOT00000018385
|
Cog6
|
component of oligomeric golgi complex 6 |
| chr2_-_123851854 | 3.38 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr15_+_83442144 | 3.33 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr7_-_116712387 | 3.19 |
ENSRNOT00000088064
|
Top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr20_+_1172867 | 3.05 |
ENSRNOT00000041189
|
Olr1708
|
olfactory receptor 1708 |
| chr5_+_60850852 | 3.00 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr4_+_154215250 | 2.80 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr4_-_70628470 | 2.80 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr3_+_94549180 | 2.59 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr16_+_23475351 | 2.43 |
ENSRNOT00000068177
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_121822781 | 2.41 |
ENSRNOT00000072082
|
Vom1r95
|
vomeronasal 1 receptor 95 |
| chr3_-_48451650 | 2.37 |
ENSRNOT00000007356
|
Gcg
|
glucagon |
| chr8_+_79054237 | 2.30 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr13_+_80125391 | 2.26 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr4_-_125929002 | 2.17 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr16_+_10250404 | 2.12 |
ENSRNOT00000087037
ENSRNOT00000081183 |
Gdf10
|
growth differentiation factor 10 |
| chr7_-_124020574 | 2.11 |
ENSRNOT00000055988
|
Poldip3
|
DNA polymerase delta interacting protein 3 |
| chr2_-_5577369 | 2.04 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_97723901 | 2.01 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr3_+_75936795 | 1.99 |
ENSRNOT00000007603
|
Olr588
|
olfactory receptor 588 |
| chrX_+_113510621 | 1.96 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr10_-_87153982 | 1.60 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr1_+_247107397 | 1.59 |
ENSRNOT00000082290
|
Cdc37l1
|
cell division cycle 37-like 1 |
| chr17_-_27451832 | 1.55 |
ENSRNOT00000084417
|
Riok1
|
RIO kinase 1 |
| chr4_+_121760773 | 1.52 |
ENSRNOT00000087069
|
Vom1r92
|
vomeronasal 1 receptor 92 |
| chr11_+_45401403 | 1.51 |
ENSRNOT00000002240
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr17_+_8135251 | 1.51 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr7_+_143897014 | 1.46 |
ENSRNOT00000017314
|
Espl1
|
extra spindle pole bodies like 1, separase |
| chr2_+_193627243 | 1.45 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chr7_+_66624512 | 1.30 |
ENSRNOT00000079410
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr16_-_79793619 | 1.27 |
ENSRNOT00000073971
|
Arhgef10
|
Rho guanine nucleotide exchange factor 10 |
| chr1_-_170175350 | 1.23 |
ENSRNOT00000023349
|
Olr201
|
olfactory receptor 201 |
| chr11_+_67451549 | 1.15 |
ENSRNOT00000042777
|
Stfa2l2
|
stefin A2-like 2 |
| chr5_+_64053946 | 1.13 |
ENSRNOT00000011622
|
Invs
|
inversin |
| chr3_+_94530586 | 1.08 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr13_-_73055631 | 1.04 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chrX_-_31759161 | 0.99 |
ENSRNOT00000041515
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr8_-_69164758 | 0.82 |
ENSRNOT00000013933
|
Map2k1
|
mitogen activated protein kinase kinase 1 |
| chr8_+_87630916 | 0.82 |
ENSRNOT00000016428
|
Myo6
|
myosin VI |
| chr13_+_29839867 | 0.79 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chrX_-_159244879 | 0.78 |
ENSRNOT00000030445
|
Map7d3
|
MAP7 domain containing 3 |
| chr1_+_230056710 | 0.74 |
ENSRNOT00000073539
|
Olr357
|
olfactory receptor 357 |
| chr2_+_150344307 | 0.74 |
ENSRNOT00000072617
|
Aadacl2
|
arylacetamide deacetylase-like 2 |
| chr11_+_66932614 | 0.66 |
ENSRNOT00000003189
|
Slc15a2
|
solute carrier family 15 member 2 |
| chr10_-_86645529 | 0.64 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr20_+_37876650 | 0.62 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr18_+_40087823 | 0.59 |
ENSRNOT00000041763
|
AABR07031947.1
|
|
| chr1_+_229480184 | 0.58 |
ENSRNOT00000044115
|
Olr387
|
olfactory receptor 387 |
| chrX_-_76925195 | 0.57 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr20_+_1124169 | 0.41 |
ENSRNOT00000072544
|
LOC100910894
|
olfactory receptor 14J1-like |
| chr7_+_17956631 | 0.40 |
ENSRNOT00000064298
|
Vom1r106
|
vomeronasal 1 receptor 106 |
| chr1_-_76722965 | 0.36 |
ENSRNOT00000052129
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr10_+_71332803 | 0.35 |
ENSRNOT00000087428
|
Synrg
|
synergin, gamma |
| chr1_+_64740487 | 0.25 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr11_-_45688560 | 0.19 |
ENSRNOT00000081508
|
Olr1536
|
olfactory receptor 1536 |
| chr11_+_43329700 | 0.11 |
ENSRNOT00000060892
|
Olr1540
|
olfactory receptor 1540 |
| chr5_-_128333805 | 0.09 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr1_+_189432604 | 0.09 |
ENSRNOT00000034610
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr1_-_77750960 | 0.06 |
ENSRNOT00000018858
|
Crx
|
cone-rod homeobox |
| chr10_+_83856280 | 0.05 |
ENSRNOT00000008964
|
Snf8
|
SNF8, ESCRT-II complex subunit |
| chr1_+_127604197 | 0.02 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 37.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 6.7 | 33.4 | GO:0061743 | motor learning(GO:0061743) |
| 4.8 | 42.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 3.0 | 15.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 2.7 | 29.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 2.5 | 14.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 2.2 | 6.7 | GO:0035723 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) regulation of monocyte extravasation(GO:2000437) |
| 2.0 | 10.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.8 | 18.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.8 | 5.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 1.7 | 6.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.7 | 6.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.6 | 4.7 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.4 | 5.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.2 | 6.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 1.2 | 4.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.0 | 5.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.8 | 3.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.8 | 12.5 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.7 | 4.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.7 | 18.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.7 | 5.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.6 | 12.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.6 | 11.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.6 | 19.6 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.6 | 2.4 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.5 | 7.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 5.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 8.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.4 | 15.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.3 | 24.6 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.3 | 1.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 0.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 1.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 31.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.8 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 6.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 3.9 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 2.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 2.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 4.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 16.4 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 3.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 21.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 10.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 3.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 7.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 7.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.5 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 1.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 7.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 14.5 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 11.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 3.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 28.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 10.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.0 | GO:0043328 | regulation of multivesicular body size(GO:0010796) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 2.5 | 24.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 2.2 | 33.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.8 | 42.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.6 | 4.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.6 | 4.7 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) gamma-tubulin small complex(GO:0008275) |
| 1.4 | 29.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.1 | 4.4 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 1.1 | 37.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 8.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 6.2 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 1.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 15.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 30.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 3.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 3.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 3.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 22.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 6.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 11.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 4.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 16.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 5.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 3.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 30.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 20.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.3 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 11.2 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 3.1 | 37.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.6 | 41.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 2.3 | 13.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 2.2 | 26.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.8 | 5.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.7 | 6.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.5 | 18.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 1.5 | 7.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.3 | 30.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.0 | 8.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.0 | 5.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.9 | 18.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.9 | 24.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.7 | 15.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.7 | 4.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.7 | 8.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.7 | 3.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.7 | 5.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.6 | 3.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.6 | 8.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.5 | 4.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 5.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.5 | 6.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 15.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 10.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 6.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 18.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 3.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 1.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 7.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 0.7 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.2 | 39.0 | GO:0016874 | ligase activity(GO:0016874) |
| 0.2 | 3.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 11.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 11.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 4.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 11.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.6 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 2.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 23.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 7.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 2.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 24.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 21.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 6.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 15.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 8.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 21.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 29.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.1 | 19.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.7 | 37.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.7 | 41.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.7 | 8.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 6.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 30.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 14.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 6.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 8.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 5.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 3.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 1.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 5.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |