Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
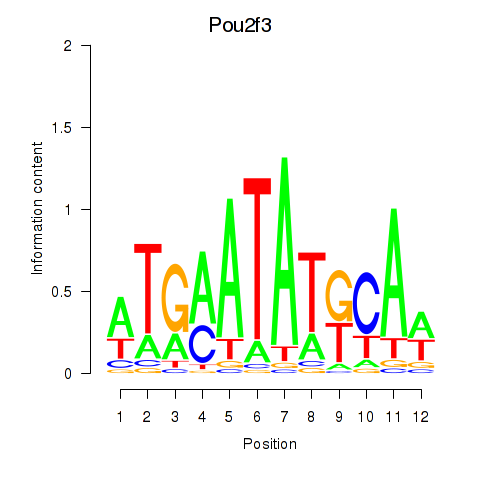
Results for Pou2f3
Z-value: 1.19
Transcription factors associated with Pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f3
|
ENSRNOG00000009118 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f3 | rn6_v1_chr8_-_47494982_47494982 | 0.16 | 4.3e-03 | Click! |
Activity profile of Pou2f3 motif
Sorted Z-values of Pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_18574909 | 37.29 |
ENSRNOT00000083090
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr8_+_39305128 | 37.24 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr1_+_27476375 | 36.80 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr11_+_66316606 | 36.18 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr11_+_42259761 | 34.48 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr16_+_78539489 | 28.42 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr16_-_39719187 | 28.19 |
ENSRNOT00000092971
|
Gpm6a
|
glycoprotein m6a |
| chr4_-_11610518 | 27.32 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_80358211 | 27.19 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr3_-_44086006 | 22.79 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr3_-_105279462 | 21.94 |
ENSRNOT00000010679
|
Scg5
|
secretogranin V |
| chr10_-_27366665 | 21.85 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr8_-_8524643 | 21.69 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr8_+_82043403 | 21.60 |
ENSRNOT00000091789
|
Myo5a
|
myosin VA |
| chr15_+_4850122 | 21.02 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr3_+_51687809 | 19.93 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr15_+_87886783 | 19.91 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr16_+_3293599 | 19.51 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr10_-_59888198 | 18.81 |
ENSRNOT00000093482
ENSRNOT00000049311 ENSRNOT00000093230 |
Aspa
|
aspartoacylase |
| chr14_+_84237520 | 18.57 |
ENSRNOT00000083580
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr3_+_58164931 | 18.49 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr19_-_11326139 | 18.34 |
ENSRNOT00000025669
|
Mt3
|
metallothionein 3 |
| chr20_-_13142856 | 18.14 |
ENSRNOT00000001743
|
S100b
|
S100 calcium binding protein B |
| chr10_+_49259194 | 17.55 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chr1_+_240355149 | 17.46 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr10_-_18574710 | 17.32 |
ENSRNOT00000079031
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr11_+_58624198 | 17.09 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr5_-_9381214 | 16.86 |
ENSRNOT00000039237
|
RGD1561849
|
similar to RIKEN cDNA 3110035E14 |
| chr4_+_24222500 | 16.24 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr7_+_133856101 | 16.01 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr10_-_66873948 | 15.92 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr1_-_24924918 | 15.80 |
ENSRNOT00000043416
|
AABR07000740.1
|
|
| chr3_-_92290919 | 15.77 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr7_-_70552897 | 15.77 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr18_-_24182012 | 15.76 |
ENSRNOT00000023594
|
Syt4
|
synaptotagmin 4 |
| chr15_-_65507968 | 15.69 |
ENSRNOT00000046240
|
Calm2
|
calmodulin 2 |
| chr7_-_13751271 | 15.21 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr10_+_1920529 | 15.09 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr9_-_88816898 | 15.04 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr7_+_44009069 | 14.92 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr7_+_28654733 | 14.62 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr17_+_23661429 | 14.58 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_+_69415057 | 14.31 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr9_-_75528644 | 14.11 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr9_+_67774150 | 14.10 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr1_+_188420461 | 13.93 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr3_-_114728410 | 13.83 |
ENSRNOT00000000181
|
Gatm
|
glycine amidinotransferase |
| chr4_+_157726941 | 13.60 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr19_+_387374 | 13.57 |
ENSRNOT00000075433
|
LOC102547811
|
uncharacterized LOC102547811 |
| chr9_-_45558993 | 13.47 |
ENSRNOT00000087866
ENSRNOT00000017714 |
Chst10
|
carbohydrate sulfotransferase 10 |
| chr10_-_56211891 | 13.00 |
ENSRNOT00000015076
|
Atp1b2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr16_+_2379480 | 12.98 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr19_-_43596801 | 12.97 |
ENSRNOT00000025625
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr1_-_101773508 | 12.93 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr2_+_3662763 | 12.92 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr4_+_136512201 | 12.91 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr18_+_57654290 | 12.87 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr18_-_31109025 | 12.51 |
ENSRNOT00000026381
|
Fchsd1
|
FCH and double SH3 domains 1 |
| chr3_-_143192408 | 12.30 |
ENSRNOT00000006885
|
LOC689081
|
similar to cystatin E2 |
| chr1_+_17602281 | 12.12 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr3_-_63094177 | 11.94 |
ENSRNOT00000045862
|
Pde11a
|
phosphodiesterase 11A |
| chr3_+_69549673 | 11.78 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr14_-_28967980 | 11.42 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr3_+_19045214 | 10.97 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr5_+_122390522 | 10.96 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_110700403 | 10.72 |
ENSRNOT00000092379
|
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr3_-_52447622 | 10.68 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr1_+_73837944 | 10.58 |
ENSRNOT00000036413
|
Lair1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr1_+_8310577 | 10.39 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr2_+_104290726 | 10.34 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr2_+_186703541 | 10.26 |
ENSRNOT00000093342
|
Fcrl1
|
Fc receptor-like 1 |
| chr6_-_142903440 | 10.21 |
ENSRNOT00000075707
|
AABR07065823.2
|
|
| chr5_-_139921013 | 10.15 |
ENSRNOT00000079957
|
Smap2
|
small ArfGAP2 |
| chr4_+_70828894 | 9.47 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr5_-_153840178 | 9.46 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr2_-_173563273 | 9.40 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr6_-_141488290 | 9.35 |
ENSRNOT00000067336
|
AABR07065792.1
|
|
| chr15_+_56666012 | 9.31 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chrX_+_35869538 | 9.28 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr7_-_8060373 | 9.18 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr14_+_34389991 | 9.17 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr6_-_142933110 | 9.12 |
ENSRNOT00000046885
|
AABR07065823.1
|
|
| chr3_-_138683318 | 8.91 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr8_+_80965255 | 8.89 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr17_+_47721977 | 8.88 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr18_+_30474947 | 8.67 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr19_+_12097632 | 8.67 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr16_-_75340360 | 8.59 |
ENSRNOT00000018501
|
Defa5
|
defensin alpha 5 |
| chr14_+_7171613 | 8.56 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr1_+_201901342 | 8.39 |
ENSRNOT00000036657
|
RGD1559891
|
similar to synaptonemal complex protein 3 |
| chr3_+_170996193 | 8.29 |
ENSRNOT00000008959
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr16_-_75481115 | 8.20 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr3_-_148312420 | 8.07 |
ENSRNOT00000047416
ENSRNOT00000081272 |
Bcl2l1
|
Bcl2-like 1 |
| chr4_+_101645731 | 8.05 |
ENSRNOT00000087901
|
AABR07060953.1
|
|
| chr3_+_117422704 | 7.95 |
ENSRNOT00000085647
|
Slc12a1
|
solute carrier family 12 member 1 |
| chr13_-_76049363 | 7.94 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr6_-_143065639 | 7.86 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chr2_+_116970344 | 7.80 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr1_-_189181901 | 7.75 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr16_+_71242470 | 7.65 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr6_-_143702033 | 7.55 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr8_+_71547468 | 7.45 |
ENSRNOT00000042369
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr14_-_20816588 | 7.43 |
ENSRNOT00000033806
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr14_+_22091777 | 7.42 |
ENSRNOT00000072483
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr3_-_148312791 | 7.42 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr9_-_65790347 | 7.29 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr15_-_29548400 | 7.26 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr18_+_29951094 | 7.21 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr4_-_117111653 | 7.18 |
ENSRNOT00000057537
|
Sfxn5
|
sideroflexin 5 |
| chr14_+_22107416 | 7.15 |
ENSRNOT00000060179
ENSRNOT00000080114 |
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr17_+_43673294 | 7.12 |
ENSRNOT00000074109
|
Hist2h4a
|
histone cluster 2, H4 |
| chr16_-_24951612 | 7.08 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr10_+_30169257 | 7.03 |
ENSRNOT00000006436
|
Rnf145
|
ring finger protein 145 |
| chrX_-_38468360 | 7.01 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chrX_+_6791136 | 7.01 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr3_-_66279155 | 6.95 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr19_+_21541742 | 6.90 |
ENSRNOT00000021095
ENSRNOT00000042077 |
Abcc12
|
ATP binding cassette subfamily C member 12 |
| chr4_-_140503412 | 6.89 |
ENSRNOT00000079266
|
AABR07061707.1
|
|
| chr3_+_101791337 | 6.89 |
ENSRNOT00000046937
|
Slc5a12
|
solute carrier family 5 member 12 |
| chr16_+_32457521 | 6.77 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr1_-_189182306 | 6.72 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr2_-_154418629 | 6.57 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr18_+_86299463 | 6.57 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr3_+_17546566 | 6.53 |
ENSRNOT00000050825
|
AABR07051583.1
|
|
| chrX_+_96991658 | 6.53 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr9_+_10941613 | 6.51 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr11_-_54344615 | 6.48 |
ENSRNOT00000090307
|
Myh15
|
myosin, heavy chain 15 |
| chr6_-_138744480 | 6.47 |
ENSRNOT00000089387
|
AABR07065651.5
|
|
| chr1_+_53531076 | 6.45 |
ENSRNOT00000018015
|
Tcp10b
|
t-complex protein 10b |
| chr13_-_74740458 | 6.44 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr6_-_61405195 | 6.44 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr2_-_154418920 | 6.40 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr4_-_69211671 | 6.32 |
ENSRNOT00000017852
|
LOC286960
|
preprotrypsinogen IV |
| chr13_+_48427038 | 6.27 |
ENSRNOT00000009241
|
Ctse
|
cathepsin E |
| chr4_+_101639641 | 6.27 |
ENSRNOT00000058282
|
AABR07060952.1
|
|
| chr4_-_176294997 | 6.14 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr6_-_142635763 | 6.10 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr9_-_63291350 | 5.97 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr2_+_196304492 | 5.96 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr11_-_61287914 | 5.95 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr1_-_67134827 | 5.80 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr1_+_199019289 | 5.75 |
ENSRNOT00000087142
|
Phkg2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr2_+_92549479 | 5.65 |
ENSRNOT00000082912
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr1_+_219403970 | 5.60 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr5_-_70463546 | 5.56 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr13_+_48426820 | 5.45 |
ENSRNOT00000048391
|
Ctse
|
cathepsin E |
| chr6_-_99273033 | 5.44 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr8_-_96547568 | 5.42 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr4_+_148139528 | 5.41 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr2_+_186776644 | 5.32 |
ENSRNOT00000046778
|
Fcrl3
|
Fc receptor-like 3 |
| chr9_-_65736852 | 5.27 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr15_-_27733785 | 5.26 |
ENSRNOT00000029488
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr6_-_142676432 | 5.26 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr17_-_43821536 | 5.24 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr4_+_56748494 | 5.14 |
ENSRNOT00000065747
|
LOC103692087
|
uncharacterized LOC103692087 |
| chr1_-_22269831 | 5.13 |
ENSRNOT00000081804
|
Stx7
|
syntaxin 7 |
| chr1_+_66959610 | 5.13 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr6_-_142372031 | 5.08 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr13_-_47623849 | 5.04 |
ENSRNOT00000006106
|
Il24
|
interleukin 24 |
| chr6_-_143131118 | 4.94 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr14_-_6679878 | 4.88 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr8_+_78872102 | 4.87 |
ENSRNOT00000090047
|
Zfp280d
|
zinc finger protein 280D |
| chr2_+_27713880 | 4.87 |
ENSRNOT00000083143
|
Gcnt4
|
glucosaminyl (N-acetyl) transferase 4, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| chr10_-_70220558 | 4.74 |
ENSRNOT00000041389
ENSRNOT00000076398 ENSRNOT00000076596 |
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr15_+_31642169 | 4.61 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chrX_-_142248369 | 4.51 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_159410799 | 4.51 |
ENSRNOT00000079225
|
Akap3
|
A-kinase anchoring protein 3 |
| chr13_-_78521911 | 4.51 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr4_+_70776046 | 4.49 |
ENSRNOT00000040403
|
Prss1
|
protease, serine 1 |
| chr10_+_28449335 | 4.48 |
ENSRNOT00000029118
|
Atp10b
|
ATPase phospholipid transporting 10B (putative) |
| chr5_-_57896475 | 4.47 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr12_+_23514981 | 4.46 |
ENSRNOT00000001937
|
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr14_-_23604834 | 4.43 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr7_+_59200918 | 4.37 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_-_104870382 | 4.34 |
ENSRNOT00000084398
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr18_+_750776 | 4.34 |
ENSRNOT00000021981
|
Zfp136
|
zinc finger protein 136 |
| chr16_-_54533871 | 4.32 |
ENSRNOT00000015144
|
LOC100910977
|
disintegrin and metalloproteinase domain-containing protein 25-like |
| chr1_+_227640680 | 4.28 |
ENSRNOT00000033613
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr14_-_34177455 | 4.27 |
ENSRNOT00000059814
ENSRNOT00000091230 |
Exoc1
|
exocyst complex component 1 |
| chrX_-_45000363 | 4.26 |
ENSRNOT00000035538
|
Cldn34e
|
claudin 34E |
| chr8_-_17696989 | 4.26 |
ENSRNOT00000083752
|
AABR07069336.1
|
|
| chr4_+_52199416 | 4.22 |
ENSRNOT00000009537
|
Spam1
|
sperm adhesion molecule 1 |
| chr4_+_88328061 | 4.20 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr4_+_87026530 | 4.17 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr13_+_88644520 | 4.17 |
ENSRNOT00000003979
|
Spata46
|
spermatogenesis associated 46 |
| chr2_+_206392200 | 4.15 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chr20_+_11114164 | 4.11 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr20_-_6195463 | 4.05 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr20_+_3677474 | 4.04 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr9_+_70059683 | 3.98 |
ENSRNOT00000016038
|
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr15_-_71779033 | 3.94 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr4_+_96449351 | 3.94 |
ENSRNOT00000091272
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr4_+_174692331 | 3.92 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr9_-_104870225 | 3.91 |
ENSRNOT00000065171
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr16_-_48921242 | 3.91 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chrX_+_136488691 | 3.88 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr18_+_30424814 | 3.82 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr6_-_10592454 | 3.67 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr6_-_138662365 | 3.64 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr4_+_102068556 | 3.64 |
ENSRNOT00000077412
|
AABR07060971.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 6.2 | 18.5 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 6.1 | 18.3 | GO:0097213 | regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 5.4 | 21.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 5.3 | 15.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 5.2 | 15.5 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 5.0 | 15.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 4.6 | 13.9 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 3.9 | 27.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 3.9 | 27.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 3.7 | 14.6 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 3.6 | 14.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 3.5 | 13.8 | GO:1990402 | embryonic liver development(GO:1990402) |
| 3.2 | 13.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 3.2 | 15.8 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 3.1 | 9.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 3.1 | 37.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.8 | 19.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 2.6 | 7.9 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 2.6 | 15.7 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 2.6 | 18.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 2.4 | 54.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 2.3 | 11.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 2.2 | 13.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 2.2 | 6.6 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.8 | 7.2 | GO:0015746 | citrate transport(GO:0015746) |
| 1.8 | 14.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.8 | 5.3 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 1.7 | 17.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.7 | 6.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.6 | 4.9 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 1.6 | 11.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.6 | 21.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.5 | 6.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 1.5 | 10.7 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 1.5 | 4.4 | GO:1904139 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.4 | 11.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.4 | 5.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 1.3 | 21.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 1.3 | 5.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.1 | 3.4 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 1.1 | 21.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 1.1 | 4.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.9 | 1.9 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.9 | 27.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.9 | 2.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.9 | 6.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.9 | 11.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.9 | 15.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.9 | 5.3 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.9 | 16.7 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.9 | 11.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.9 | 17.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.9 | 3.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.8 | 11.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.8 | 2.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.8 | 10.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.8 | 4.7 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.8 | 34.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.7 | 28.4 | GO:0001964 | startle response(GO:0001964) |
| 0.7 | 12.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.7 | 2.2 | GO:2000196 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of female gonad development(GO:2000196) negative regulation of eosinophil migration(GO:2000417) |
| 0.7 | 9.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.7 | 2.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 5.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.6 | 8.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.6 | 4.5 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 12.9 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.6 | 1.9 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.6 | 6.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.6 | 33.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.6 | 13.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.6 | 15.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.6 | 2.9 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.6 | 2.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 4.9 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.5 | 2.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 20.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 3.1 | GO:0002503 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.5 | 1.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 8.5 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.4 | 3.9 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.4 | 8.6 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.4 | 13.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.4 | 1.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 33.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.4 | 2.9 | GO:0050957 | equilibrioception(GO:0050957) righting reflex(GO:0060013) |
| 0.4 | 1.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 12.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 4.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.3 | 1.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 42.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.3 | 2.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.3 | 2.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 3.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.3 | 2.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 2.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.3 | 6.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.7 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 | 7.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.9 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 3.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 2.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.2 | 4.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 30.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 18.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 2.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 6.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 14.6 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.2 | 1.8 | GO:0042501 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.2 | 4.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.7 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 2.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 6.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.5 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.2 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 4.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 5.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 7.0 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 3.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 1.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 2.6 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 10.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 3.0 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.1 | 13.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 9.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.9 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 4.5 | GO:0003014 | renal system process(GO:0003014) |
| 0.1 | 8.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 1.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 11.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 4.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.7 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 3.2 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 8.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 3.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.4 | GO:0007283 | spermatogenesis(GO:0007283) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.6 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 4.5 | 13.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 3.8 | 22.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.7 | 27.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 2.3 | 18.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 2.2 | 15.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 2.1 | 17.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.5 | 30.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.4 | 4.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.2 | 15.8 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.2 | 4.7 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 1.1 | 15.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.1 | 13.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.0 | 21.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.8 | 5.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.7 | 11.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.7 | 28.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.6 | 1.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 9.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.6 | 1.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.5 | 56.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 53.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.5 | 6.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 2.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 2.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 12.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 3.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 2.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 4.5 | GO:0002177 | manchette(GO:0002177) |
| 0.4 | 4.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.4 | 21.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 6.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 6.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 10.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 3.4 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 0.9 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.3 | 14.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 16.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 7.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 7.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 19.4 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 2.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 14.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 10.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 40.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 22.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 6.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 9.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 17.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 20.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 11.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 8.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 57.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 8.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 18.5 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 4.0 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 6.3 | 18.8 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 5.2 | 15.7 | GO:0031997 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 5.0 | 15.0 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 4.6 | 18.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 4.4 | 21.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 4.3 | 13.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 4.3 | 12.9 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 4.3 | 17.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 3.2 | 15.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.1 | 9.3 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 2.6 | 7.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 2.6 | 15.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 2.4 | 14.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.3 | 6.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.1 | 8.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.0 | 18.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.9 | 21.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.8 | 7.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 1.7 | 34.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 1.7 | 11.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.7 | 18.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.6 | 14.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.5 | 6.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 1.5 | 4.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.4 | 7.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.4 | 11.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 1.3 | 19.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.2 | 20.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.1 | 37.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 1.0 | 15.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.0 | 15.2 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.9 | 13.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.9 | 2.7 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.9 | 6.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 3.4 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.8 | 2.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.8 | 50.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.7 | 17.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.7 | 9.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.6 | 1.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 9.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.6 | 10.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.6 | 10.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.6 | 2.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.6 | 1.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.6 | 2.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.5 | 5.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.5 | 6.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 10.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 1.9 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.5 | 13.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 49.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.4 | 12.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 17.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.4 | 14.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 7.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.4 | 1.8 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.3 | 3.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 6.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 5.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 2.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 2.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 2.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 4.1 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.3 | 6.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 13.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 3.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.4 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.2 | 21.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 0.7 | GO:0050145 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 4.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 4.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 6.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 14.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 6.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 2.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 3.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 2.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 4.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 11.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 1.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 4.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 19.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 1.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 9.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 20.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 22.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 7.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 4.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.9 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 7.4 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 9.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 6.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 5.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 40.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 16.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 11.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 11.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 26.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.7 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 21.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.0 | 15.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.9 | 34.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.7 | 14.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 17.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 7.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 4.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 4.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 18.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 6.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 8.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 6.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 12.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 22.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.3 | 15.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.3 | 18.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.3 | 21.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 18.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.1 | 27.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 1.0 | 12.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.0 | 27.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 1.0 | 8.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.9 | 21.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.8 | 14.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.8 | 15.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.8 | 13.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.7 | 19.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 15.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 14.1 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.5 | 14.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 13.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 10.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 2.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 7.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 16.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 2.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 14.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 2.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 8.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 12.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 32.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 6.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 4.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 2.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 6.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |