Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
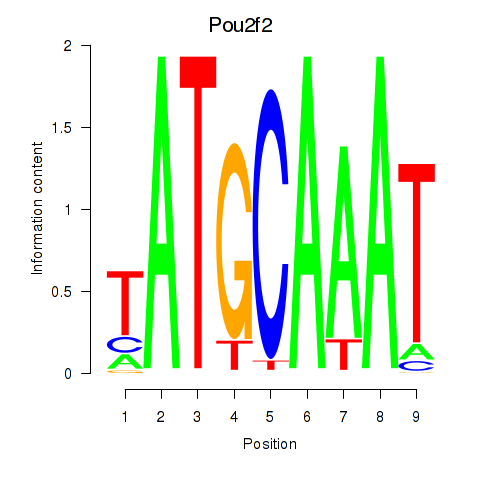
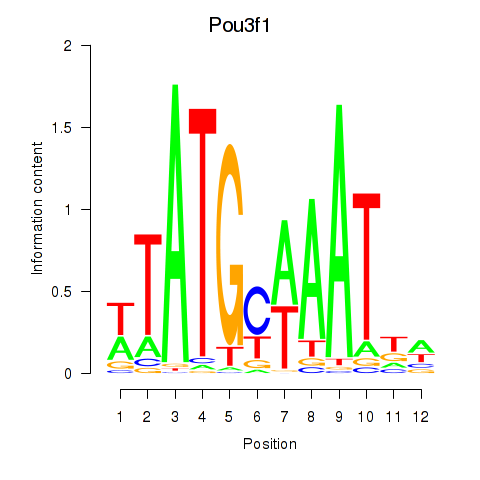
Results for Pou2f2_Pou3f1
Z-value: 4.08
Transcription factors associated with Pou2f2_Pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f2
|
ENSRNOG00000055650 | POU class 2 homeobox 2 |
|
Pou3f1
|
ENSRNOG00000047686 | Pou3f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f2 | rn6_v1_chr1_-_82004538_82004538 | 0.74 | 4.3e-56 | Click! |
Activity profile of Pou2f2_Pou3f1 motif
Sorted Z-values of Pou2f2_Pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_18315320 | 287.36 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr1_-_17378047 | 276.30 |
ENSRNOT00000020102
|
Themis
|
thymocyte selection associated |
| chr4_+_102489916 | 275.88 |
ENSRNOT00000082031
|
AABR07061001.1
|
|
| chr6_-_138662365 | 275.41 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr6_-_140102325 | 265.44 |
ENSRNOT00000072238
|
AABR07065750.2
|
|
| chr11_+_85532526 | 250.28 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr3_+_19274273 | 241.34 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr4_-_103258134 | 235.87 |
ENSRNOT00000086827
|
AABR07061052.1
|
|
| chr7_-_107634287 | 229.31 |
ENSRNOT00000093672
ENSRNOT00000087116 |
Sla
|
src-like adaptor |
| chr4_+_106323089 | 229.30 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr3_+_17889972 | 226.53 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr6_-_138632159 | 225.96 |
ENSRNOT00000082921
ENSRNOT00000040702 |
Ighm
|
immunoglobulin heavy constant mu |
| chr11_+_85508300 | 213.35 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr6_+_139405966 | 205.68 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr6_-_140642221 | 199.79 |
ENSRNOT00000081996
|
AABR07065772.2
|
|
| chr6_-_142635763 | 199.58 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr4_+_98370797 | 195.93 |
ENSRNOT00000031991
|
AABR07060872.1
|
|
| chr11_+_85618714 | 195.25 |
ENSRNOT00000074614
|
AC109901.1
|
|
| chr6_+_139428999 | 194.13 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chr4_-_102124609 | 193.07 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr6_-_140880070 | 191.58 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr2_+_223121410 | 189.09 |
ENSRNOT00000087559
|
AABR07013116.1
|
|
| chr7_-_18793289 | 185.42 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr6_-_143131118 | 181.93 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr6_-_142353308 | 178.90 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr6_-_138736203 | 178.74 |
ENSRNOT00000052021
|
LOC100360169
|
rCG21044-like |
| chr11_+_85042348 | 170.00 |
ENSRNOT00000042220
|
AABR07034718.1
|
|
| chr17_+_43632397 | 162.15 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr17_+_43423111 | 161.52 |
ENSRNOT00000022630
|
Hist1h2ba
|
histone cluster 1 H2B family member a |
| chr3_+_16571602 | 160.75 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr4_+_102665529 | 160.56 |
ENSRNOT00000082333
|
AABR07061005.1
|
|
| chr17_+_44520537 | 160.46 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr10_-_94500591 | 155.34 |
ENSRNOT00000015976
|
Cd79b
|
CD79b molecule |
| chr11_+_85992696 | 153.70 |
ENSRNOT00000084433
|
AABR07034736.1
|
|
| chr6_-_141008427 | 153.48 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chr3_+_17546566 | 152.55 |
ENSRNOT00000050825
|
AABR07051583.1
|
|
| chr6_-_138954577 | 148.59 |
ENSRNOT00000042728
|
AABR07065656.7
|
|
| chr10_-_45297385 | 147.49 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr6_-_138954741 | 146.21 |
ENSRNOT00000083278
|
AABR07065656.7
|
|
| chr4_+_102068556 | 146.00 |
ENSRNOT00000077412
|
AABR07060971.1
|
|
| chr17_-_44520240 | 144.31 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr10_+_45289741 | 144.15 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr17_+_44528125 | 142.03 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr6_-_138931429 | 137.58 |
ENSRNOT00000090584
|
LOC100359993
|
Ighg protein-like |
| chr2_+_198418691 | 136.09 |
ENSRNOT00000089409
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr6_-_143065639 | 133.61 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chr6_-_139660666 | 132.47 |
ENSRNOT00000086098
|
AABR07065705.1
|
|
| chr3_+_18706988 | 132.38 |
ENSRNOT00000074650
|
AABR07051652.1
|
|
| chr6_-_141198565 | 132.33 |
ENSRNOT00000064361
|
AABR07065782.1
|
|
| chr17_-_43422846 | 132.07 |
ENSRNOT00000050526
|
Hist1h2aa
|
histone cluster 1 H2A family member a |
| chr6_-_139004980 | 131.91 |
ENSRNOT00000085427
ENSRNOT00000087351 |
AABR07065656.4
|
|
| chr6_-_142060032 | 129.99 |
ENSRNOT00000064717
|
AABR07065811.1
|
|
| chr6_-_138631997 | 126.51 |
ENSRNOT00000073304
|
AABR07065651.3
|
|
| chr17_-_44527801 | 125.04 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr3_+_20303979 | 124.25 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr4_+_98481520 | 123.08 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chrX_+_28593405 | 122.35 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr17_+_44758460 | 121.72 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr2_-_198382190 | 120.50 |
ENSRNOT00000044268
|
Hist2h2aa2
|
histone cluster 2, H2aa2 |
| chr2_+_198388809 | 119.94 |
ENSRNOT00000083087
|
Hist2h2aa3
|
histone cluster 2, H2aa3 |
| chr6_-_139637187 | 113.58 |
ENSRNOT00000089454
|
LOC100359993
|
Ighg protein-like |
| chrX_+_96991658 | 112.84 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr11_+_85561460 | 111.50 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr6_-_139637354 | 111.19 |
ENSRNOT00000072900
|
LOC100359993
|
Ighg protein-like |
| chr6_-_143702033 | 110.91 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr1_+_192233910 | 106.59 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr17_-_44748188 | 105.26 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr4_+_101909389 | 102.85 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chr17_+_44738643 | 102.80 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr3_+_20375699 | 101.80 |
ENSRNOT00000088492
|
AABR07051731.1
|
|
| chr6_+_139560028 | 100.63 |
ENSRNOT00000072633
|
LOC100360581
|
rCG58847-like |
| chr3_-_19320915 | 99.39 |
ENSRNOT00000043673
|
RGD1565617
|
similar to Ig variable region, light chain |
| chr19_+_3325893 | 98.88 |
ENSRNOT00000048879
|
RGD1565617
|
similar to Ig variable region, light chain |
| chr3_-_16441030 | 98.86 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr3_-_20419417 | 98.41 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr6_-_142933110 | 97.29 |
ENSRNOT00000046885
|
AABR07065823.1
|
|
| chr3_-_20479999 | 97.12 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr6_-_139033819 | 97.08 |
ENSRNOT00000091961
|
AABR07065656.6
|
|
| chr6_-_141321108 | 96.24 |
ENSRNOT00000040556
|
AABR07065789.3
|
|
| chr17_-_44738330 | 95.54 |
ENSRNOT00000072195
|
LOC100364835
|
histone cluster 1, H2bd-like |
| chr6_-_142585188 | 94.78 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr6_-_141062581 | 90.83 |
ENSRNOT00000073446
|
AABR07065778.3
|
|
| chr17_+_44748482 | 90.81 |
ENSRNOT00000083765
|
Hist1h2bl
|
histone cluster 1 H2B family member l |
| chr3_-_20695952 | 90.62 |
ENSRNOT00000072306
|
AABR07051746.1
|
|
| chr2_-_198360678 | 90.38 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr18_+_44737154 | 90.29 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr6_+_139531330 | 89.78 |
ENSRNOT00000083025
|
AABR07065699.1
|
|
| chr17_+_43627930 | 89.24 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr4_+_93888502 | 87.19 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr4_+_101639641 | 87.15 |
ENSRNOT00000058282
|
AABR07060952.1
|
|
| chr6_-_142676432 | 85.55 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr3_+_17180411 | 85.27 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr4_+_102147211 | 83.50 |
ENSRNOT00000083239
|
AABR07060980.1
|
|
| chr6_-_139710905 | 83.00 |
ENSRNOT00000077430
|
AABR07065705.2
|
|
| chr3_+_16846412 | 82.20 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr19_+_50045020 | 81.78 |
ENSRNOT00000090165
|
Plcg2
|
phospholipase C, gamma 2 |
| chr17_-_44758170 | 81.35 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr6_-_138685656 | 80.98 |
ENSRNOT00000041706
|
AABR07065651.7
|
|
| chr2_+_264704769 | 80.28 |
ENSRNOT00000012667
|
Depdc1
|
DEP domain containing 1 |
| chr17_-_43627629 | 79.67 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr2_+_55835151 | 78.60 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr6_-_141831632 | 77.60 |
ENSRNOT00000075526
|
AABR07065802.1
|
|
| chr3_-_20457554 | 76.70 |
ENSRNOT00000074237
|
AABR07051733.1
|
|
| chr3_+_19045214 | 76.33 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chrX_+_96863891 | 74.00 |
ENSRNOT00000085665
|
AABR07040284.1
|
|
| chr4_-_95970666 | 73.97 |
ENSRNOT00000008826
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_-_143206772 | 72.94 |
ENSRNOT00000073713
|
AABR07065839.1
|
|
| chr5_+_16526058 | 68.82 |
ENSRNOT00000011130
|
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr1_-_118826977 | 68.48 |
ENSRNOT00000079274
|
AABR07003833.1
|
|
| chr6_+_139293294 | 67.99 |
ENSRNOT00000050297
ENSRNOT00000081817 |
LOC100359993
|
Ighg protein-like |
| chr6_-_142880382 | 66.78 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr6_-_141957537 | 65.50 |
ENSRNOT00000090358
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr4_-_103761881 | 63.62 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr1_-_265798167 | 58.50 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr4_-_119327822 | 58.47 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr4_-_28437676 | 56.75 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr2_+_198359754 | 55.58 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr3_+_17139670 | 55.32 |
ENSRNOT00000073316
|
AABR07051564.1
|
|
| chr3_-_92749121 | 55.00 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chrX_-_62690806 | 53.27 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr11_+_85633243 | 53.12 |
ENSRNOT00000045807
|
LOC682352
|
Ig lambda chain V-VI region AR-like |
| chr17_-_43689311 | 52.37 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr6_-_141472746 | 48.30 |
ENSRNOT00000048010
|
AABR07065792.2
|
|
| chr2_+_198360998 | 47.55 |
ENSRNOT00000046129
|
Hist2h2be
|
histone cluster 2, H2be |
| chr6_-_143768985 | 47.52 |
ENSRNOT00000046694
|
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr12_+_38160464 | 46.77 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr6_-_141365198 | 46.14 |
ENSRNOT00000040523
|
AABR07065789.2
|
|
| chr10_-_19652898 | 45.97 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr6_-_55524436 | 44.05 |
ENSRNOT00000006752
|
Tspan13
|
tetraspanin 13 |
| chr9_-_54457753 | 43.17 |
ENSRNOT00000020032
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr3_+_79918969 | 39.74 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr3_+_19071980 | 39.53 |
ENSRNOT00000079487
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr6_-_125723732 | 38.70 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr4_+_93959152 | 38.63 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr5_-_12172009 | 38.32 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr20_+_44680449 | 38.24 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr2_-_250778269 | 37.95 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr4_+_147333056 | 37.39 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr10_-_108196217 | 37.34 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr6_-_141866756 | 37.15 |
ENSRNOT00000068561
|
AABR07065804.1
|
|
| chr4_-_183424449 | 36.43 |
ENSRNOT00000071930
|
Fam60a
|
family with sequence similarity 60, member A |
| chr1_-_142183884 | 35.54 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr6_-_142178771 | 34.68 |
ENSRNOT00000071577
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr6_-_125723944 | 33.27 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr14_-_86078954 | 32.51 |
ENSRNOT00000018319
|
Polm
|
DNA polymerase mu |
| chr1_-_57327379 | 31.70 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr18_+_12008759 | 31.22 |
ENSRNOT00000022665
ENSRNOT00000061344 ENSRNOT00000038635 |
Dsg1
|
desmoglein 1 |
| chr16_-_21017163 | 31.19 |
ENSRNOT00000027661
|
Mef2b
|
myocyte enhancer factor 2B |
| chr11_-_33003021 | 30.15 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr16_-_22561496 | 29.78 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
| chr6_-_140587251 | 28.32 |
ENSRNOT00000090704
|
AABR07065772.4
|
|
| chr17_+_23116661 | 27.94 |
ENSRNOT00000067374
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_+_102290338 | 27.31 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr10_-_13898855 | 27.04 |
ENSRNOT00000004249
|
Rab26
|
RAB26, member RAS oncogene family |
| chr17_-_31916553 | 26.35 |
ENSRNOT00000074220
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr4_-_183426439 | 25.94 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chrX_+_104684676 | 25.77 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr10_-_50402616 | 25.41 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr10_+_95642640 | 25.07 |
ENSRNOT00000029782
|
RGD1359290
|
Ribosomal_L22 domain containing protein RGD1359290 |
| chr18_-_12640716 | 25.07 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr10_-_78993045 | 24.17 |
ENSRNOT00000043005
|
LOC100363469
|
ribosomal protein S24-like |
| chr3_-_94833406 | 24.14 |
ENSRNOT00000049695
|
RGD1560017
|
similar to Ac2-210 |
| chr1_-_179010257 | 24.00 |
ENSRNOT00000017199
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr4_+_155359921 | 23.86 |
ENSRNOT00000020762
|
Aicda
|
activation-induced cytidine deaminase |
| chr20_-_22459025 | 22.83 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr6_-_143537030 | 22.10 |
ENSRNOT00000071876
|
LOC100360610
|
rCG64220-like |
| chr6_-_36876805 | 21.71 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr3_-_90751055 | 20.97 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr2_-_139528162 | 19.85 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr17_-_15519060 | 19.54 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr3_-_20518105 | 19.01 |
ENSRNOT00000050435
|
AABR07051735.1
|
|
| chr11_-_60679555 | 18.98 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_-_18244535 | 18.50 |
ENSRNOT00000040689
|
AABR07051626.1
|
|
| chr3_+_2396143 | 18.19 |
ENSRNOT00000012388
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr6_-_92527711 | 17.93 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr20_-_13657943 | 16.82 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr16_-_7412150 | 16.40 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr2_+_187893368 | 16.19 |
ENSRNOT00000092031
|
Mex3a
|
mex-3 RNA binding family member A |
| chr18_-_70924708 | 16.04 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr17_+_15749978 | 15.47 |
ENSRNOT00000067311
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_141715846 | 15.44 |
ENSRNOT00000044966
|
AABR07065798.1
|
|
| chrX_+_158835811 | 14.82 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_+_144080614 | 12.32 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr14_-_59735450 | 11.67 |
ENSRNOT00000071929
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_163112301 | 11.67 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr1_+_273854248 | 11.17 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr1_-_264172729 | 10.21 |
ENSRNOT00000018447
|
Scd
|
stearoyl-CoA desaturase |
| chr20_+_5646097 | 9.82 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr3_-_3951025 | 9.81 |
ENSRNOT00000026212
|
Notch1
|
notch 1 |
| chr3_-_164964702 | 9.72 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chr6_-_141384305 | 9.50 |
ENSRNOT00000045320
|
AABR07065790.1
|
|
| chr5_+_157434481 | 9.43 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr15_+_12827707 | 8.79 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr9_-_119321500 | 8.57 |
ENSRNOT00000048125
|
Myl12b
|
myosin light chain 12B |
| chr9_+_43049587 | 7.56 |
ENSRNOT00000021434
ENSRNOT00000079234 |
Cnnm4
|
cyclin and CBS domain divalent metal cation transport mediator 4 |
| chr20_+_9173127 | 7.42 |
ENSRNOT00000088409
|
AABR07044520.1
|
|
| chr14_+_86514214 | 7.27 |
ENSRNOT00000052002
|
LOC103693776
|
zinc finger MIZ domain-containing protein 2 |
| chr8_+_12823155 | 6.85 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chr12_+_25264192 | 6.84 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr2_+_182723854 | 6.73 |
ENSRNOT00000012658
|
Sfrp2
|
secreted frizzled-related protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f2_Pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.5 | 106.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 26.9 | 161.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 22.9 | 68.8 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 18.5 | 74.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 17.5 | 122.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 17.1 | 308.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 14.6 | 58.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 14.5 | 276.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 14.5 | 101.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 14.5 | 43.4 | GO:0060846 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) blood vessel endothelial cell fate specification(GO:0097101) |
| 13.3 | 53.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 13.0 | 1274.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 12.5 | 37.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 10.8 | 43.2 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 10.0 | 30.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 9.9 | 39.7 | GO:0045347 | hypermethylation of CpG island(GO:0044027) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 8.2 | 81.8 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 7.6 | 22.8 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 7.2 | 72.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 6.4 | 598.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 5.3 | 16.0 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 4.9 | 9.8 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 4.3 | 25.8 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 4.3 | 29.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 3.6 | 25.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 3.5 | 257.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 3.4 | 10.2 | GO:1903699 | tarsal gland development(GO:1903699) |
| 3.1 | 46.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 2.7 | 2.7 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 2.7 | 32.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 2.7 | 106.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 2.2 | 6.7 | GO:2000041 | sclerotome development(GO:0061056) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 2.2 | 8.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.1 | 38.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 2.0 | 18.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 2.0 | 9.8 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.8 | 17.9 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 1.6 | 1.6 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 1.4 | 9.7 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 1.4 | 12.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.2 | 82.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 1.2 | 19.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.1 | 6.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.1 | 78.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 1.1 | 7.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.0 | 12.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.8 | 4.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.7 | 33.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.7 | 0.7 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.7 | 4.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 5.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 24.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.5 | 7.6 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.4 | 31.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.4 | 19.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.4 | 27.0 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.3 | 2.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 5.6 | GO:0060746 | parental behavior(GO:0060746) |
| 0.2 | 1.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 38.3 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.2 | 16.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 6.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 3.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 19.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 2.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 13.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.8 | 155.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 24.4 | 2388.5 | GO:0000786 | nucleosome(GO:0000786) |
| 22.9 | 68.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 18.3 | 55.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 14.4 | 72.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 7.1 | 276.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 6.7 | 53.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 5.4 | 21.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 4.2 | 62.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 3.3 | 46.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.6 | 17.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 2.5 | 37.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 2.3 | 29.8 | GO:0042627 | chylomicron(GO:0042627) |
| 1.6 | 227.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 1.4 | 31.2 | GO:0030057 | desmosome(GO:0030057) |
| 1.2 | 106.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 1.0 | 23.9 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.9 | 19.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.8 | 6.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.7 | 59.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.5 | 25.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.5 | 19.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 8.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.4 | 9.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 37.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.4 | 50.3 | GO:0030496 | midbody(GO:0030496) |
| 0.3 | 24.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 30.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 79.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 1.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 3.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 19.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 11.2 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 25.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 8.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 6.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.5 | 106.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 9.6 | 38.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 7.6 | 68.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 7.6 | 53.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 7.3 | 58.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 6.8 | 27.0 | GO:0019002 | GMP binding(GO:0019002) |
| 6.6 | 19.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 5.5 | 55.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 5.3 | 37.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 5.1 | 46.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 5.0 | 39.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 4.2 | 25.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 4.2 | 257.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 4.0 | 23.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 3.9 | 43.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 3.5 | 31.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 3.3 | 69.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 2.9 | 90.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 2.7 | 16.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 2.6 | 31.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 2.6 | 10.2 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.5 | 1679.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 2.5 | 12.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.5 | 9.8 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 2.2 | 46.8 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 2.1 | 37.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 2.1 | 74.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.5 | 32.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.5 | 44.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.2 | 8.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 22.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.0 | 6.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.9 | 31.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.9 | 6.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.8 | 19.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.8 | 5.9 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.7 | 19.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 58.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.5 | 2.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.5 | 9.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.5 | 7.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 9.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 9.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 11.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 49.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 15.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 31.4 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 15.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 106.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 4.0 | 68.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 3.6 | 43.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 3.4 | 237.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 3.3 | 153.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 3.1 | 55.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.1 | 122.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 1.8 | 32.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.6 | 78.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 1.3 | 149.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 1.2 | 37.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.9 | 32.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.5 | 9.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.5 | 30.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.5 | 39.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 4.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 52.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 4.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 6.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.0 | 1144.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 8.1 | 315.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 5.1 | 71.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 4.2 | 55.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 3.6 | 43.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 3.1 | 53.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 3.0 | 56.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 2.6 | 31.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 2.2 | 31.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.9 | 35.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 1.8 | 29.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.3 | 11.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 1.3 | 122.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.9 | 25.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.6 | 6.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 80.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.5 | 19.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 8.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 12.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 6.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.3 | 2.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 46.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |