Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
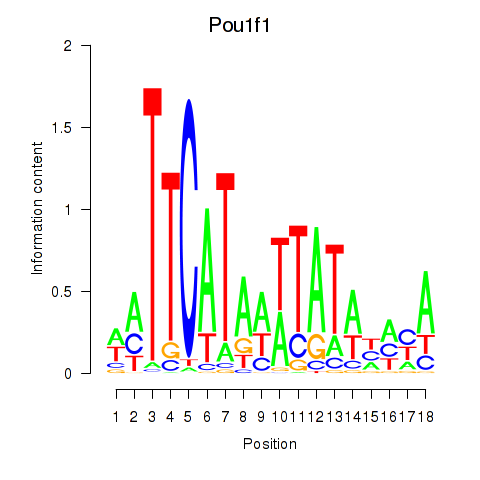
Results for Pou1f1
Z-value: 2.14
Transcription factors associated with Pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou1f1
|
ENSRNOG00000000715 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou1f1 | rn6_v1_chr11_+_2645865_2645865 | -0.48 | 7.1e-20 | Click! |
Activity profile of Pou1f1 motif
Sorted Z-values of Pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22724399 | 60.66 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr1_-_148119857 | 56.89 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr1_+_147713892 | 46.49 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr6_-_127816055 | 42.70 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr3_-_63836017 | 42.12 |
ENSRNOT00000030978
|
AABR07052585.1
|
|
| chr1_-_76722965 | 41.28 |
ENSRNOT00000052129
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr10_+_53781239 | 40.07 |
ENSRNOT00000082871
|
Myh2
|
myosin heavy chain 2 |
| chr1_+_83163079 | 37.99 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr1_-_76614279 | 37.97 |
ENSRNOT00000041367
ENSRNOT00000089371 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr17_+_32973695 | 36.31 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr14_+_22806132 | 35.99 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr14_+_22517774 | 35.91 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr2_-_216443518 | 34.83 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr16_+_54153054 | 33.24 |
ENSRNOT00000090644
ENSRNOT00000014248 |
Fgl1
|
fibrinogen-like 1 |
| chr11_+_7265828 | 31.62 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr4_+_109497962 | 31.41 |
ENSRNOT00000057869
|
Reg1a
|
regenerating family member 1 alpha |
| chr4_-_129619142 | 31.27 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr14_-_115052450 | 29.81 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr5_-_77408323 | 29.69 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr1_-_76517134 | 29.02 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr10_+_96639924 | 28.01 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr14_+_22251499 | 26.91 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr3_-_37803112 | 26.63 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr13_-_56958549 | 26.06 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chr4_-_41212072 | 25.77 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr13_-_56693968 | 24.64 |
ENSRNOT00000060160
|
AABR07021096.1
|
|
| chr4_-_70747226 | 23.71 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr2_-_216348194 | 23.47 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr2_-_248484100 | 23.36 |
ENSRNOT00000040856
|
Gbp3
|
guanylate binding protein 3 |
| chr14_+_22375955 | 23.21 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr5_-_77316764 | 23.06 |
ENSRNOT00000071395
ENSRNOT00000076464 |
Mup4
|
major urinary protein 4 |
| chr5_+_90338795 | 22.85 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr7_-_138483612 | 22.65 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_-_77342299 | 22.59 |
ENSRNOT00000075994
|
RGD1566134
|
similar to alpha-2u-globulin |
| chr7_+_2689501 | 22.53 |
ENSRNOT00000041341
|
Apof
|
apolipoprotein F |
| chr1_-_258766881 | 22.41 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr1_+_148240504 | 21.43 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr19_+_27404712 | 21.27 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_+_70614524 | 21.21 |
ENSRNOT00000041100
|
Prss3
|
protease, serine 3 |
| chr1_+_83744238 | 21.20 |
ENSRNOT00000028249
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr17_+_33028293 | 20.46 |
ENSRNOT00000072764
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr1_-_275882444 | 20.08 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr18_+_56193978 | 19.56 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_+_52753011 | 19.44 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr6_+_127946686 | 19.41 |
ENSRNOT00000082680
|
LOC500712
|
Ab1-233 |
| chr2_-_227207584 | 19.20 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chrX_-_110232179 | 19.19 |
ENSRNOT00000014739
|
Serpina7
|
serpin family A member 7 |
| chr1_-_258877045 | 18.97 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr4_-_70628470 | 18.96 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr1_+_229030233 | 18.70 |
ENSRNOT00000084503
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr14_+_26662965 | 18.67 |
ENSRNOT00000002621
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr6_-_47890033 | 18.59 |
ENSRNOT00000011290
|
Colec11
|
collectin sub-family member 11 |
| chr18_-_15540177 | 18.47 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr20_+_26999795 | 18.38 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr5_-_19368431 | 18.34 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr17_+_72160735 | 18.18 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr2_+_74360622 | 18.04 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr1_+_248723397 | 17.84 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr2_+_88217188 | 17.77 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr4_+_14109864 | 17.58 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr6_-_128003418 | 17.55 |
ENSRNOT00000013896
|
Serpina3c
|
serine (or cysteine) proteinase inhibitor, clade A, member 3C |
| chr6_+_128048099 | 17.52 |
ENSRNOT00000084685
ENSRNOT00000087017 |
LOC500712
|
Ab1-233 |
| chr2_+_147496229 | 17.45 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr7_-_14435967 | 17.24 |
ENSRNOT00000074801
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_+_106896790 | 17.08 |
ENSRNOT00000042952
ENSRNOT00000064193 |
Ano5
|
anoctamin 5 |
| chrX_+_110233170 | 16.76 |
ENSRNOT00000036019
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chr2_+_227080924 | 16.72 |
ENSRNOT00000029871
|
Fabp2
|
fatty acid binding protein 2 |
| chr9_-_4327679 | 16.69 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr1_-_76780230 | 16.66 |
ENSRNOT00000002046
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr20_-_28263037 | 16.48 |
ENSRNOT00000030348
|
Edar
|
ectodysplasin-A receptor |
| chr4_-_13878126 | 16.47 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chr4_+_70755795 | 16.38 |
ENSRNOT00000043527
|
LOC683849
|
similar to Anionic trypsin II precursor (Pretrypsinogen II) |
| chr13_-_56763981 | 16.23 |
ENSRNOT00000087916
|
LOC100361907
|
complement factor H-related protein B |
| chr15_-_23969011 | 15.72 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr4_+_70689737 | 15.67 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr6_-_23291568 | 15.59 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr14_+_22072024 | 15.57 |
ENSRNOT00000002680
|
ste2
|
estrogen sulfotransferase |
| chr10_+_70262361 | 15.48 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr1_+_137860946 | 15.39 |
ENSRNOT00000031334
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr18_+_35574002 | 15.38 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr3_-_7141522 | 15.35 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr19_-_41686229 | 15.16 |
ENSRNOT00000022721
|
Tat
|
tyrosine aminotransferase |
| chr5_-_77492013 | 15.10 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chrX_-_13601069 | 15.07 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr14_-_1791636 | 15.05 |
ENSRNOT00000072859
|
Vom2r73
|
vomeronasal 2 receptor, 73 |
| chr9_+_37749059 | 15.00 |
ENSRNOT00000071970
|
LOC100909868
|
myotilin-like |
| chr1_+_248428099 | 14.90 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr15_-_34550850 | 14.88 |
ENSRNOT00000027794
ENSRNOT00000090228 |
Cbln3
|
cerebellin 3 precursor |
| chr4_+_154391647 | 14.85 |
ENSRNOT00000081488
ENSRNOT00000079192 |
Mug1
|
murinoglobulin 1 |
| chrM_+_7758 | 14.85 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr1_-_116919269 | 14.81 |
ENSRNOT00000035149
|
AABR07003616.1
|
|
| chr1_+_107262659 | 14.67 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr10_+_53713938 | 14.63 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr2_-_173668555 | 14.60 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr3_+_54253949 | 14.27 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr9_-_79530821 | 14.25 |
ENSRNOT00000085619
|
Mreg
|
melanoregulin |
| chr18_-_62476700 | 14.09 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr8_-_111777602 | 13.97 |
ENSRNOT00000052317
ENSRNOT00000083855 |
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr7_-_29171783 | 13.96 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr7_+_38858062 | 13.90 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr1_-_258875572 | 13.90 |
ENSRNOT00000093005
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr9_+_74124016 | 13.84 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr3_-_74815509 | 13.82 |
ENSRNOT00000045173
|
Olr499
|
olfactory receptor 499 |
| chr14_+_22724070 | 13.79 |
ENSRNOT00000089471
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr8_-_77398156 | 13.79 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr4_+_166833320 | 13.68 |
ENSRNOT00000031438
|
Tas2r120
|
taste receptor, type 2, member 120 |
| chr2_-_211017778 | 13.66 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr17_-_69827112 | 13.65 |
ENSRNOT00000023835
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr16_+_25773602 | 13.61 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr2_+_54466280 | 13.41 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr9_+_119353840 | 13.37 |
ENSRNOT00000085362
|
Myom1
|
myomesin 1 |
| chr6_-_138507135 | 13.31 |
ENSRNOT00000071078
|
AABR07065640.1
|
|
| chr14_-_19132208 | 13.24 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr16_+_39909270 | 13.19 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr14_-_62595854 | 13.19 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr5_+_165465095 | 13.16 |
ENSRNOT00000031978
|
LOC691354
|
hypothetical protein LOC691354 |
| chr8_+_58431407 | 13.15 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr1_+_65576535 | 13.09 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr19_+_14835822 | 12.97 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr2_+_22950018 | 12.95 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr15_-_49505553 | 12.90 |
ENSRNOT00000028974
|
Adamdec1
|
ADAM-like, decysin 1 |
| chrM_+_11736 | 12.84 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_+_154215250 | 12.68 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr14_+_84282073 | 12.57 |
ENSRNOT00000078271
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr13_+_91481461 | 12.53 |
ENSRNOT00000012103
|
Olr1576
|
olfactory receptor 1576 |
| chr7_-_3805023 | 12.51 |
ENSRNOT00000065917
|
Olr881
|
olfactory receptor 881 |
| chr19_+_15294248 | 12.47 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr8_+_52729003 | 12.42 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr4_-_176231344 | 12.33 |
ENSRNOT00000049154
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr14_-_1785698 | 12.29 |
ENSRNOT00000071182
|
Vom2r73
|
vomeronasal 2 receptor, 73 |
| chr6_+_128046780 | 12.19 |
ENSRNOT00000078064
|
LOC500712
|
Ab1-233 |
| chrX_+_51286737 | 12.16 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr6_-_23316962 | 12.13 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_7210209 | 11.94 |
ENSRNOT00000076695
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr14_-_773281 | 11.87 |
ENSRNOT00000045104
ENSRNOT00000050676 ENSRNOT00000049917 |
Vom2r-ps125
|
vomeronasal 2 receptor, pseudogene 125 |
| chr2_-_19808937 | 11.86 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr4_-_176026133 | 11.82 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr2_+_243502073 | 11.80 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_-_42430928 | 11.80 |
ENSRNOT00000072172
|
Olr1257
|
olfactory receptor 1257 |
| chr14_+_812382 | 11.77 |
ENSRNOT00000041506
ENSRNOT00000039993 |
AABR07014089.1
|
|
| chr8_-_38154344 | 11.72 |
ENSRNOT00000068043
|
LOC100360095
|
urinary protein 1-like |
| chr5_-_22769907 | 11.72 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr16_-_75518336 | 11.72 |
ENSRNOT00000058037
|
Defa11
|
defensin alpha 11 |
| chr16_+_50181316 | 11.70 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr7_+_11769400 | 11.63 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr4_+_14212925 | 11.54 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr3_-_127500709 | 11.53 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chrX_-_104337203 | 11.52 |
ENSRNOT00000050903
|
AABR07040565.1
|
|
| chr4_+_122244711 | 11.44 |
ENSRNOT00000038251
|
Uroc1
|
urocanate hydratase 1 |
| chr9_+_8054466 | 11.40 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr3_-_52447622 | 11.37 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr7_+_139685573 | 11.31 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr13_-_52834134 | 11.31 |
ENSRNOT00000038874
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr4_+_166076156 | 11.26 |
ENSRNOT00000007124
|
Prp2
|
proline rich protein 2 |
| chr17_+_15966234 | 11.16 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr2_-_243407608 | 11.09 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr10_-_46206135 | 11.08 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr4_+_103495993 | 11.07 |
ENSRNOT00000072325
|
AABR07061068.1
|
|
| chr18_-_64061472 | 11.01 |
ENSRNOT00000022519
|
Fam210a
|
family with sequence similarity 210, member A |
| chr3_-_74790822 | 11.00 |
ENSRNOT00000045888
|
LOC100912505
|
olfactory receptor 8K3-like |
| chr8_+_87211819 | 11.00 |
ENSRNOT00000086093
|
LOC100363289
|
LRRGT00022-like |
| chr3_+_75936795 | 10.91 |
ENSRNOT00000007603
|
Olr588
|
olfactory receptor 588 |
| chr1_+_229722768 | 10.86 |
ENSRNOT00000017126
|
Olr339
|
olfactory receptor 339 |
| chr9_+_94286550 | 10.82 |
ENSRNOT00000026504
|
Chrnd
|
cholinergic receptor nicotinic delta subunit |
| chr11_+_42259761 | 10.77 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr9_-_17693200 | 10.68 |
ENSRNOT00000026716
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chrX_+_15669191 | 10.64 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chr3_+_69549673 | 10.53 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr9_-_50762082 | 10.47 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr1_+_229889771 | 10.43 |
ENSRNOT00000085941
|
Olr348
|
olfactory receptor 348 |
| chr1_+_94718402 | 10.40 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chrX_-_56765893 | 10.40 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr8_-_38549268 | 10.39 |
ENSRNOT00000088001
|
LOC100912026
|
urinary protein 3-like |
| chr19_-_22194740 | 10.36 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_+_116869957 | 10.35 |
ENSRNOT00000075203
|
Vom2r25
|
vomeronasal 2 receptor, 25 |
| chr11_-_43617633 | 10.30 |
ENSRNOT00000060874
|
Olr1557
|
olfactory receptor 1557 |
| chr3_+_112228919 | 10.23 |
ENSRNOT00000011761
|
Capn3
|
calpain 3 |
| chr6_-_127656603 | 10.22 |
ENSRNOT00000015516
|
Serpina11
|
serpin family A member 11 |
| chr2_+_22910236 | 10.15 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chrX_+_145558840 | 10.14 |
ENSRNOT00000032522
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chr1_-_49844547 | 10.14 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr3_+_112228720 | 10.12 |
ENSRNOT00000079079
|
Capn3
|
calpain 3 |
| chr3_+_11653529 | 10.11 |
ENSRNOT00000091048
|
Ak1
|
adenylate kinase 1 |
| chr11_+_20474483 | 10.10 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr16_-_40025401 | 10.05 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr4_+_101639641 | 10.03 |
ENSRNOT00000058282
|
AABR07060952.1
|
|
| chr3_-_78016906 | 10.01 |
ENSRNOT00000089048
|
Olr687
|
olfactory receptor 687 |
| chrX_-_64726210 | 9.99 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr11_-_81444375 | 9.87 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr7_-_70552897 | 9.87 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr5_-_77433847 | 9.86 |
ENSRNOT00000076906
ENSRNOT00000043056 |
LOC500473
Mup4
|
similar to alpha-2u globulin PGCL2 major urinary protein 4 |
| chrX_-_123867149 | 9.83 |
ENSRNOT00000089754
|
Rhox3
|
reproductive homeobox on X chromosome 3 |
| chr1_-_173456488 | 9.83 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chrX_-_65400298 | 9.79 |
ENSRNOT00000032121
|
Vsig4
|
V-set and immunoglobulin domain containing 4 |
| chr20_-_1109027 | 9.78 |
ENSRNOT00000051315
|
Olr1696
|
olfactory receptor 1696 |
| chr5_+_132005738 | 9.77 |
ENSRNOT00000041421
|
Skint4
|
selection and upkeep of intraepithelial T cells 4 |
| chr4_+_146276862 | 9.69 |
ENSRNOT00000009705
|
Slc6a1
|
solute carrier family 6 member 1 |
| chr1_-_67853552 | 9.68 |
ENSRNOT00000080755
ENSRNOT00000003545 |
Vom2r32
|
vomeronasal 2 receptor, 32 |
| chr16_+_74292438 | 9.65 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr4_+_175814118 | 9.64 |
ENSRNOT00000013409
ENSRNOT00000013514 |
Slco1b2
|
solute carrier organic anion transporter family, member 1B2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 31.4 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 9.2 | 248.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 7.3 | 58.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 7.1 | 28.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 6.8 | 20.3 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 5.2 | 15.7 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 5.0 | 15.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 4.6 | 13.8 | GO:0010034 | response to acetate(GO:0010034) |
| 4.5 | 31.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 4.3 | 17.2 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 4.3 | 21.4 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 4.3 | 68.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 4.1 | 16.3 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 4.1 | 12.2 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 4.0 | 19.9 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 3.8 | 18.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 3.6 | 25.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 3.5 | 14.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 3.4 | 37.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 3.4 | 10.1 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 3.3 | 13.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 3.3 | 9.9 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 3.3 | 13.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 3.1 | 3.1 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 3.1 | 45.8 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 3.1 | 9.2 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 2.9 | 46.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 2.9 | 11.4 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 2.8 | 11.3 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 2.7 | 16.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 2.7 | 8.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 2.7 | 10.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 2.7 | 8.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 2.5 | 15.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.4 | 33.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 2.4 | 7.1 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 2.3 | 11.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.3 | 7.0 | GO:0045819 | cellular response to water deprivation(GO:0042631) positive regulation of glycogen catabolic process(GO:0045819) |
| 2.3 | 6.9 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 2.3 | 9.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.2 | 20.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 2.2 | 6.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 2.2 | 15.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 2.2 | 4.3 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 2.2 | 8.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 2.1 | 8.6 | GO:1903413 | negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 2.1 | 6.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.1 | 14.8 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 2.1 | 2.1 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 2.1 | 6.2 | GO:0042369 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) response to sodium phosphate(GO:1904383) |
| 2.0 | 6.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 2.0 | 12.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 2.0 | 26.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 2.0 | 6.0 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 2.0 | 17.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.9 | 5.8 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 1.9 | 9.7 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 1.9 | 5.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.9 | 20.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.9 | 49.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.8 | 27.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.8 | 5.4 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 1.8 | 5.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 1.7 | 2190.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 1.7 | 3.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.7 | 8.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 1.7 | 5.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 1.7 | 26.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 1.7 | 1.7 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 1.6 | 6.6 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.6 | 4.9 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.6 | 6.5 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 1.6 | 6.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.6 | 8.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.6 | 9.5 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 1.5 | 3.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 1.5 | 6.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.5 | 9.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.5 | 7.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.5 | 6.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 1.5 | 11.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.4 | 4.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.4 | 10.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.4 | 5.8 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 1.4 | 5.8 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 1.4 | 5.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.4 | 8.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.4 | 5.6 | GO:1903576 | response to L-arginine(GO:1903576) |
| 1.4 | 18.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.4 | 5.4 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 1.4 | 6.8 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 1.3 | 4.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.3 | 4.0 | GO:0046687 | response to chromate(GO:0046687) |
| 1.3 | 12.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.3 | 21.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 1.3 | 9.2 | GO:0015669 | gas transport(GO:0015669) |
| 1.3 | 10.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.3 | 3.8 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.2 | 2.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.2 | 6.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 1.2 | 4.9 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 1.2 | 27.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.2 | 8.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.2 | 2.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.1 | 3.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 1.1 | 13.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.1 | 12.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.1 | 3.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.1 | 6.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 1.1 | 3.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.1 | 2.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.1 | 4.4 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 1.0 | 5.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 1.0 | 3.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.0 | 10.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.0 | 18.5 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 1.0 | 1.0 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 1.0 | 2.9 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 1.0 | 2.9 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.9 | 5.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.9 | 13.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.9 | 2.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.9 | 3.7 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.9 | 2.7 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.9 | 2.7 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.9 | 2.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.9 | 5.2 | GO:0010266 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 0.9 | 3.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.9 | 5.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.8 | 16.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.8 | 2.5 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.8 | 2.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.8 | 8.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.8 | 8.0 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.8 | 8.0 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.8 | 2.4 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 0.8 | 6.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.8 | 8.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.8 | 3.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.8 | 13.1 | GO:0009750 | response to fructose(GO:0009750) |
| 0.8 | 3.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.8 | 5.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.8 | 3.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.8 | 8.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.7 | 40.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.7 | 2.2 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.7 | 9.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.7 | 5.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.7 | 5.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.7 | 2.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.7 | 3.7 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.7 | 11.5 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.7 | 0.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.7 | 8.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 2.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.7 | 8.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.7 | 10.8 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.7 | 3.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.7 | 11.4 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.7 | 6.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.7 | 4.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 13.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.6 | 1.9 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.6 | 32.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.6 | 3.0 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.6 | 6.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.6 | 1.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.6 | 17.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.6 | 1.8 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 | 2.9 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.6 | 9.8 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.6 | 2.8 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.5 | 4.9 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.5 | 3.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 136.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.5 | 10.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.5 | 2.7 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.5 | 2.7 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 0.5 | 4.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.5 | 2.7 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 2.7 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 9.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.5 | 2.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 3.6 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.5 | 4.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.5 | 4.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.5 | 8.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.5 | 1.5 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.5 | 3.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 4.7 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.5 | 0.5 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.5 | 10.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.5 | 1.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.5 | 7.7 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.5 | 4.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.4 | 6.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 4.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.4 | 11.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 4.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.4 | 21.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.4 | 4.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 0.4 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.4 | 18.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.4 | 1.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 1.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 1.6 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.4 | 1.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.4 | 2.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 4.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.4 | 1.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 0.8 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.4 | 5.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.4 | 1.1 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.4 | 1.9 | GO:0009624 | response to nematode(GO:0009624) |
| 0.4 | 5.9 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.4 | 2.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 5.2 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.4 | 3.7 | GO:1990834 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) response to odorant(GO:1990834) |
| 0.4 | 0.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.4 | 1.8 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.4 | 1.1 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.4 | 2.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 | 2.5 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.4 | 0.7 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 | 1.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 1.4 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.4 | 10.6 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.3 | 1.7 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.3 | 6.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 1.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.3 | 1.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 0.7 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.3 | 2.0 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.3 | 25.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.3 | 2.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 3.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 5.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.3 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 3.8 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.3 | 1.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 10.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 7.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 5.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 3.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 3.9 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.3 | 1.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 3.8 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.3 | 1.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 1.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.3 | 5.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.3 | 3.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.3 | 1.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 1.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 4.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 1.4 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.3 | 3.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.3 | 1.9 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.3 | 1.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 2.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 1.8 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.3 | 4.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 3.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 1.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 1.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 1.8 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.3 | 2.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 2.4 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 4.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 1.9 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 0.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 1.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 2.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 2.9 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 3.8 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 2.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 2.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 12.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.6 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 7.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 2.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 2.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 2.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 2.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 4.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.2 | 2.7 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.2 | 1.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 1.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.7 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.4 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.2 | 1.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 1.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 2.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 2.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 2.6 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.2 | 3.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 2.9 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 7.0 | GO:0051339 | regulation of lyase activity(GO:0051339) |
| 0.2 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 8.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 2.8 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 3.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 4.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 1.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 3.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 5.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 4.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.8 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 2.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.8 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 1.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 2.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.3 | GO:1903999 | regulation of eating behavior(GO:1903998) negative regulation of eating behavior(GO:1903999) |
| 0.1 | 4.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 1.4 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 4.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 5.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 7.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.8 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 2.2 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 1.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 1.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 2.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.8 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.6 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.1 | 2.0 | GO:0043039 | tRNA aminoacylation for protein translation(GO:0006418) tRNA aminoacylation(GO:0043039) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.4 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 2.8 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 1.0 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 54.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 4.9 | 19.6 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 3.8 | 11.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 3.1 | 9.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 2.9 | 29.5 | GO:0045179 | apical cortex(GO:0045179) |
| 2.9 | 14.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 2.6 | 29.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 2.5 | 27.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.3 | 68.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 2.3 | 9.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.2 | 11.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 2.0 | 8.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.5 | 6.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.4 | 4.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 1.3 | 19.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 1.3 | 147.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.3 | 3.8 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 1.2 | 63.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.2 | 8.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 6.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.2 | 14.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.1 | 8.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.1 | 35.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 1.1 | 11.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.1 | 14.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 1.0 | 3.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.0 | 3.1 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 1.0 | 9.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.9 | 4.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.9 | 11.9 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.9 | 2.7 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.9 | 39.4 | GO:0043034 | costamere(GO:0043034) |
| 0.9 | 5.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.8 | 6.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.8 | 9.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.8 | 0.8 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.8 | 4.5 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.7 | 82.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.7 | 3.7 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.7 | 28.3 | GO:0031672 | A band(GO:0031672) |
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 19.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.7 | 13.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 7.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 3.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 3.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 4.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 1.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.6 | 2.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.6 | 4.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 13.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 1.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.6 | 1.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.5 | 10.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 1.6 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.5 | 8.8 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.5 | 2.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.5 | 1.5 | GO:0000802 | transverse filament(GO:0000802) |
| 0.5 | 3.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.5 | 12.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.5 | 1.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 2.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 8.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 3.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 3.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 11.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 4.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 25.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 22.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 8.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 6.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 3.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 1.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.4 | 3.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 61.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.4 | 1.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.4 | 2.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 44.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 8.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 2.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 1.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 2.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.3 | 13.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 5.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 2.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.3 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 7.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 2.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 2.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 5.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 5.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1925.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.3 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 2.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 12.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 3.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 26.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 249.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 8.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 1.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 5.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 4.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 3.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 2.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 2.0 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 5.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 1.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.7 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 9.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 4.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 3.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 2.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 33.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.4 | 58.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 17.0 | 85.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 14.5 | 43.6 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 9.3 | 28.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 8.2 | 247.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 6.4 | 199.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 5.3 | 21.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 5.1 | 15.4 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 5.1 | 15.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 4.6 | 23.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 4.4 | 13.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 4.3 | 17.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 4.0 | 15.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 3.8 | 19.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.8 | 19.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 3.8 | 22.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.7 | 29.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 3.4 | 13.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 3.4 | 10.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 3.3 | 20.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 3.0 | 3.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 3.0 | 14.9 | GO:0005534 | galactose binding(GO:0005534) |
| 3.0 | 17.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 2.9 | 41.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 2.8 | 16.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 2.4 | 17.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.4 | 19.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.4 | 11.8 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 2.3 | 37.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 2.3 | 7.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 2.3 | 9.2 | GO:0019002 | GMP binding(GO:0019002) |
| 2.2 | 15.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 2.2 | 6.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 2.2 | 6.7 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 2.2 | 8.9 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 2.2 | 13.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 2.2 | 8.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 2.1 | 6.4 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 2.1 | 6.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 2.1 | 831.2 | GO:0005549 | odorant binding(GO:0005549) |
| 2.0 | 8.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 2.0 | 6.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 2.0 | 13.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 2.0 | 27.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 2.0 | 19.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 2.0 | 7.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.8 | 11.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.8 | 5.4 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.7 | 5.2 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 1.7 | 5.1 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 1.7 | 15.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 1.6 | 11.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 1.6 | 9.9 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.6 | 34.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.6 | 19.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.6 | 4.9 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.6 | 8.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.6 | 42.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.6 | 18.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.5 | 4.6 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 1.5 | 1350.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 1.5 | 5.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.5 | 8.9 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.5 | 1.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.5 | 10.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.5 | 5.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.4 | 17.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 1.4 | 17.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.4 | 25.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.4 | 16.7 | GO:0031432 | titin binding(GO:0031432) |
| 1.4 | 6.9 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 1.3 | 5.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.3 | 8.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.3 | 1.3 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) alkane 1-monooxygenase activity(GO:0018685) |
| 1.3 | 5.2 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.3 | 6.4 | GO:0019862 | IgA binding(GO:0019862) |
| 1.3 | 3.8 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.3 | 6.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.3 | 17.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.3 | 3.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.3 | 2.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 1.3 | 3.8 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.3 | 11.3 | GO:0070061 | fructose binding(GO:0070061) |
| 1.2 | 6.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.2 | 3.7 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 1.2 | 8.5 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 4.8 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 1.2 | 24.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.2 | 5.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.2 | 130.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.2 | 5.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 1.1 | 20.4 | GO:0001848 | complement binding(GO:0001848) |
| 1.1 | 5.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.1 | 3.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.1 | 4.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.1 | 13.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 1.1 | 3.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.0 | 12.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.0 | 2.9 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.9 | 10.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.9 | 4.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.9 | 2.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.9 | 4.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.9 | 2.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.9 | 2.7 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.9 | 2.7 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.9 | 4.4 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.9 | 1.8 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.9 | 7.0 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.9 | 3.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.9 | 6.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.8 | 6.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.8 | 2.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.8 | 16.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.8 | 8.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.8 | 2.4 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.8 | 8.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.8 | 7.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.8 | 5.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.7 | 2.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.7 | 2.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 2.9 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.7 | 10.8 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.7 | 2.9 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.7 | 2.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.7 | 5.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.7 | 2.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.7 | 6.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.6 | 4.5 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.6 | 10.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.6 | 2.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.6 | 4.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.6 | 9.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 16.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.6 | 7.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.6 | 27.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.6 | 16.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.6 | 7.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 68.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.6 | 11.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.6 | 4.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 4.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 2.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.6 | 6.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.6 | 5.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.5 | 3.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 3.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 10.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.5 | 2.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.5 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 10.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 3.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 5.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 5.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 2.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.5 | 26.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.5 | 3.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.5 | 7.6 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.5 | 2.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.5 | 3.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 5.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.5 | 2.9 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 1.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.5 | 1.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.5 | 6.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 1.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.5 | 1.8 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 4.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 4.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.4 | 3.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 91.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 6.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 15.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 4.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 4.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.4 | 7.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.4 | 7.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 1.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.4 | 1.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.4 | 3.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 1.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 3.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 1.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.4 | 23.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.4 | 7.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 2.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 2.0 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.3 | 2.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 7.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 0.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 3.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 26.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 6.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.3 | 1.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 2.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 5.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 4.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.3 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 6.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 6.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 2.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 3.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 6.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 9.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.2 | 10.9 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.2 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.9 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 4.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 5.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.6 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.2 | 2.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 12.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 0.6 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.2 | 3.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.8 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 3.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 2.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 4.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.5 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 2.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 22.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.2 | 2.4 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.2 | 4.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.2 | 3.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.9 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 4.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.9 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 8.5 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 9.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 41.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 2.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 0.9 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 2.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 14.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 10.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 31.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 2.0 | 2.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 1.4 | 2.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 1.1 | 53.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.9 | 23.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.9 | 27.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.7 | 37.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.6 | 36.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.5 | 4.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.4 | 106.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 5.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.4 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 24.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 8.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 3.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 6.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 3.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 8.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 4.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 4.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 2.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 3.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 2.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 47.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 7.6 | 143.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.4 | 21.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 2.3 | 34.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.7 | 18.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.6 | 32.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.5 | 11.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.5 | 24.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.4 | 24.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.4 | 19.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.4 | 52.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 86.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 1.0 | 8.1 | REACTOME OPSINS | Genes involved in Opsins |
| 1.0 | 12.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.9 | 2.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.9 | 10.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.8 | 61.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.7 | 10.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 19.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.7 | 6.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.7 | 11.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 12.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 15.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 18.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.6 | 103.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 2.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 7.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 5.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 6.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.5 | 8.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 2.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 11.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 1.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 12.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 5.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.4 | 2.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 10.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 18.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 3.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 11.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 16.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 5.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 11.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 5.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 4.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 3.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 4.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 5.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 6.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 7.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 4.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 1.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 18.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 16.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 8.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 3.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |