Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Plagl1
Z-value: 0.61
Transcription factors associated with Plagl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plagl1
|
ENSRNOG00000025587 | PLAG1 like zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | rn6_v1_chr1_+_7252349_7252349 | 0.28 | 3.3e-07 | Click! |
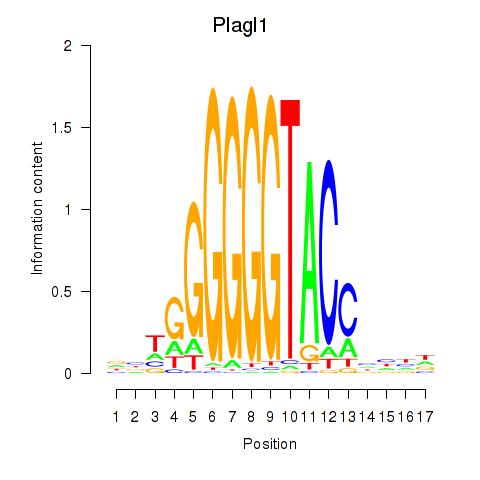
Activity profile of Plagl1 motif
Sorted Z-values of Plagl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_104095179 | 23.19 |
ENSRNOT00000093487
|
Cldn10
|
claudin 10 |
| chr12_-_29743705 | 14.66 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr10_-_31359699 | 14.13 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_12640716 | 12.59 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr6_-_24985716 | 11.74 |
ENSRNOT00000010613
|
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr10_-_88266210 | 11.74 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr1_+_219964429 | 10.79 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr8_-_57255263 | 9.90 |
ENSRNOT00000028972
|
LOC100125362
|
hypothetical protein LOC100125362 |
| chrX_-_11164915 | 9.34 |
ENSRNOT00000005138
|
Atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr5_+_133221139 | 9.17 |
ENSRNOT00000047522
|
Trabd2b
|
TraB domain containing 2B |
| chr19_+_56220755 | 9.09 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr1_-_82409639 | 8.68 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr12_-_23661009 | 8.66 |
ENSRNOT00000059451
|
Upk3bl
|
uroplakin 3B-like |
| chr12_-_36555694 | 8.58 |
ENSRNOT00000001292
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr12_+_22026075 | 7.59 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr19_+_52086325 | 7.45 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr16_+_21050243 | 6.73 |
ENSRNOT00000064308
|
Ncan
|
neurocan |
| chr1_-_227175096 | 6.67 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr4_-_72143748 | 6.60 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr14_-_83062302 | 6.36 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr3_-_160301552 | 6.24 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr6_+_30038777 | 6.18 |
ENSRNOT00000072340
|
Fam228b
|
family with sequence similarity 228, member B |
| chr10_-_55783489 | 6.05 |
ENSRNOT00000010415
|
Alox15b
|
arachidonate 15-lipoxygenase, type B |
| chr11_+_47188495 | 5.92 |
ENSRNOT00000002188
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr10_+_59533480 | 5.86 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr10_+_59529785 | 5.75 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr12_+_23151180 | 5.71 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr12_+_23463157 | 5.63 |
ENSRNOT00000044841
|
Cux1
|
cut-like homeobox 1 |
| chr16_+_80729959 | 5.58 |
ENSRNOT00000082049
|
Tdrp
|
testis development related protein |
| chr7_+_144078496 | 5.49 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_+_228544418 | 5.43 |
ENSRNOT00000013030
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr10_-_66020682 | 5.36 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr10_+_11100917 | 5.29 |
ENSRNOT00000006067
|
Coro7
|
coronin 7 |
| chr1_-_141451075 | 5.22 |
ENSRNOT00000033491
|
Kif7
|
kinesin family member 7 |
| chr13_+_90184569 | 5.19 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr5_+_154522119 | 5.19 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr7_-_12598183 | 5.11 |
ENSRNOT00000089009
|
Arid3a
|
AT-rich interaction domain 3A |
| chr1_+_7252349 | 4.95 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr17_+_9596957 | 4.95 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr1_-_222189604 | 4.86 |
ENSRNOT00000028704
|
Kcnk4
|
potassium two pore domain channel subfamily K member 4 |
| chr5_-_170679315 | 4.79 |
ENSRNOT00000071459
|
Ajap1
|
adherens junctions associated protein 1 |
| chr10_-_105368242 | 4.70 |
ENSRNOT00000075293
ENSRNOT00000072230 |
Rnf157
|
ring finger protein 157 |
| chr7_+_11095468 | 4.62 |
ENSRNOT00000061219
|
Celf5
|
CUGBP, Elav-like family member 5 |
| chr4_+_57034675 | 4.53 |
ENSRNOT00000080223
|
Smo
|
smoothened, frizzled class receptor |
| chr20_+_11114164 | 4.52 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr20_-_18060661 | 4.38 |
ENSRNOT00000070925
|
AABR07044711.1
|
|
| chr4_-_144416116 | 4.28 |
ENSRNOT00000007724
|
Oxtr
|
oxytocin receptor |
| chr13_-_85443976 | 4.16 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chrX_-_157312028 | 4.12 |
ENSRNOT00000077979
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr2_+_165486910 | 3.97 |
ENSRNOT00000012982
|
LOC100362176
|
hypothetical protein LOC100362176 |
| chr10_-_103816287 | 3.79 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr8_+_129201669 | 3.73 |
ENSRNOT00000025663
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr4_-_145147397 | 3.63 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr12_-_9990284 | 3.50 |
ENSRNOT00000001264
|
Rasl11a
|
RAS-like family 11 member A |
| chr5_+_140870140 | 3.31 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr1_+_175445088 | 3.26 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr7_-_144936803 | 3.12 |
ENSRNOT00000055279
|
Gpr84
|
G protein-coupled receptor 84 |
| chr12_+_49626871 | 3.06 |
ENSRNOT00000082593
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr8_+_71514281 | 3.02 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr8_+_107882219 | 3.01 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_12598370 | 2.98 |
ENSRNOT00000026708
|
Arid3a
|
AT-rich interaction domain 3A |
| chr4_-_85192834 | 2.79 |
ENSRNOT00000043752
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr13_-_52256196 | 2.73 |
ENSRNOT00000011105
|
Ipo9
|
importin 9 |
| chr5_+_137546860 | 2.61 |
ENSRNOT00000074431
|
Olr858
|
olfactory receptor 858 |
| chr1_-_101236065 | 2.58 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr10_-_45534570 | 2.52 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chrX_+_68774699 | 2.44 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr10_-_5253336 | 2.42 |
ENSRNOT00000085310
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr20_+_6288267 | 2.40 |
ENSRNOT00000000627
|
Srsf3
|
serine and arginine rich splicing factor 3 |
| chr4_+_121899292 | 2.37 |
ENSRNOT00000077949
|
Vom1r97
|
vomeronasal 1 receptor 97 |
| chr2_+_120266933 | 2.37 |
ENSRNOT00000015192
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr1_+_170212817 | 2.34 |
ENSRNOT00000040672
|
RGD1561034
|
similar to hypothetical protein MGC34805 |
| chrX_+_122938009 | 2.30 |
ENSRNOT00000089266
ENSRNOT00000017550 |
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_+_160852164 | 2.29 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr10_+_106785077 | 2.28 |
ENSRNOT00000075047
|
Tmc8
|
transmembrane channel-like 8 |
| chr9_+_113699170 | 2.23 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr7_-_116920507 | 2.12 |
ENSRNOT00000048363
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr7_+_15422479 | 2.10 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr1_-_88826302 | 2.07 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr8_-_132027006 | 2.03 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr1_-_213987053 | 1.88 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr15_+_30550202 | 1.87 |
ENSRNOT00000087199
|
AABR07017745.3
|
|
| chr15_+_47455690 | 1.80 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr8_-_96088364 | 1.76 |
ENSRNOT00000086161
ENSRNOT00000056818 |
Snx14
|
sorting nexin 14 |
| chr20_+_14578605 | 1.75 |
ENSRNOT00000041165
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr10_+_15108072 | 1.73 |
ENSRNOT00000060286
|
Mslnl
|
mesothelin-like |
| chr12_-_21760292 | 1.71 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr20_+_11436267 | 1.69 |
ENSRNOT00000001631
|
Trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr9_+_92618550 | 1.68 |
ENSRNOT00000078987
|
Sp140
|
SP140 nuclear body protein |
| chr16_+_41079444 | 1.56 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr5_+_14415606 | 1.53 |
ENSRNOT00000089273
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr7_-_12918173 | 1.35 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr15_-_56970365 | 1.29 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr7_+_130326600 | 1.26 |
ENSRNOT00000084776
ENSRNOT00000055808 |
Ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr18_+_29191731 | 1.22 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr15_-_33766438 | 1.22 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr13_-_50509916 | 1.21 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr15_+_34167945 | 1.18 |
ENSRNOT00000032252
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr2_-_219558675 | 1.16 |
ENSRNOT00000020149
|
Rtcd1
|
RNA terminal phosphate cyclase domain 1 |
| chr10_-_64862268 | 1.14 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr1_-_205630073 | 1.12 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr7_-_11018160 | 1.11 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr4_+_21862313 | 1.11 |
ENSRNOT00000007948
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr10_+_86657285 | 1.08 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr12_+_18996591 | 1.02 |
ENSRNOT00000011065
|
Vamp7
|
vesicle-associated membrane protein 7 |
| chr13_+_48745860 | 0.95 |
ENSRNOT00000010242
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr9_-_15306465 | 0.95 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr3_+_103866144 | 0.94 |
ENSRNOT00000007688
|
NEWGENE_1582771
|
katanin regulatory subunit B1 like 1 |
| chr2_+_252090669 | 0.92 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr3_+_172154754 | 0.86 |
ENSRNOT00000088286
|
Stx16
|
syntaxin 16 |
| chr4_+_78240385 | 0.86 |
ENSRNOT00000011041
|
Zfp775
|
zinc finger protein 775 |
| chr10_+_47065850 | 0.74 |
ENSRNOT00000077868
ENSRNOT00000066250 |
Dhrs7b
|
dehydrogenase/reductase 7B |
| chr9_+_82571269 | 0.70 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr15_+_49010492 | 0.67 |
ENSRNOT00000024527
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr1_+_212578450 | 0.63 |
ENSRNOT00000064501
|
Paox
|
polyamine oxidase |
| chr15_+_33755478 | 0.62 |
ENSRNOT00000034073
|
Thtpa
|
thiamine triphosphatase |
| chr1_+_31700953 | 0.55 |
ENSRNOT00000020251
|
Exoc3
|
exocyst complex component 3 |
| chr6_-_138068648 | 0.53 |
ENSRNOT00000081908
|
Ighm
|
immunoglobulin heavy constant mu |
| chr9_-_9702306 | 0.46 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr8_-_21523540 | 0.43 |
ENSRNOT00000085060
|
Zfp266
|
zinc finger protein 266 |
| chr3_-_10694649 | 0.42 |
ENSRNOT00000037742
|
Gpr107
|
G protein-coupled receptor 107 |
| chr3_+_122285243 | 0.34 |
ENSRNOT00000040366
|
Rpl22l2
|
ribosomal protein L22-like 2 |
| chr18_+_32594958 | 0.32 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr1_+_220806473 | 0.26 |
ENSRNOT00000027869
|
Bles03
|
basophilic leukemia expressed protein BLES03 |
| chr5_+_14415841 | 0.26 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr13_+_88644520 | 0.23 |
ENSRNOT00000003979
|
Spata46
|
spermatogenesis associated 46 |
| chr20_+_41083317 | 0.21 |
ENSRNOT00000000660
|
Tspyl1
|
TSPY-like 1 |
| chr10_+_56187020 | 0.15 |
ENSRNOT00000046490
|
Tp53
|
tumor protein p53 |
| chr4_-_157230647 | 0.12 |
ENSRNOT00000017334
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr14_-_86796378 | 0.08 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr4_-_100483168 | 0.01 |
ENSRNOT00000019713
|
Tgoln2
|
trans-golgi network protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plagl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 2.1 | 6.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.1 | 10.5 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.8 | 14.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.7 | 5.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.6 | 4.9 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 1.3 | 3.8 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.2 | 6.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.1 | 4.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.0 | 4.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.0 | 3.1 | GO:0042441 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.9 | 6.6 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.9 | 9.2 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.9 | 10.8 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.9 | 5.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.9 | 8.6 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.9 | 4.3 | GO:0034059 | positive regulation of norepinephrine secretion(GO:0010701) response to anoxia(GO:0034059) |
| 0.6 | 2.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.6 | 5.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.5 | 11.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.5 | 4.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 6.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 5.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 1.7 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.4 | 3.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.4 | 2.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 | 1.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 2.4 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 1.0 | GO:1903778 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 0.3 | 3.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.3 | 0.9 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 | 5.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 1.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 1.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 6.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 5.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 3.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.1 | 2.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 1.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.7 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 5.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 11.7 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 2.9 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 7.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 4.0 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 1.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 2.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 4.0 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.9 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 6.9 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.0 | 8.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 4.9 | GO:0007283 | spermatogenesis(GO:0007283) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 14.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 4.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.3 | 3.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.3 | 11.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 12.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 11.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 11.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 3.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 23.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 1.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 3.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 8.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.9 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 4.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 9.2 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 2.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 9.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 9.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 9.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 9.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 3.7 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.6 | 4.9 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 1.1 | 5.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 1.0 | 8.6 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.8 | 4.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.7 | 15.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 2.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.5 | 3.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 4.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 3.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 4.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 3.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 5.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 0.9 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 4.5 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.3 | 1.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.3 | 11.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 6.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 2.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 6.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 3.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 1.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 19.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 6.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 3.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 12.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 5.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 11.7 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 23.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 3.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 7.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 8.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 5.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 14.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 6.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 6.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 4.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 3.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 4.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 23.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 15.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.5 | 9.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 6.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 11.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 4.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 10.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 11.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 3.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 4.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 5.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 4.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 7.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 5.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |