Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
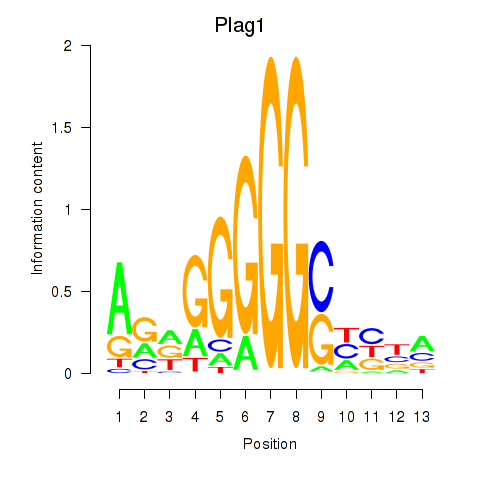
Results for Plag1
Z-value: 1.28
Transcription factors associated with Plag1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plag1
|
ENSRNOG00000008846 | PLAG1 zinc finger |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plag1 | rn6_v1_chr5_-_16799776_16799776 | 0.20 | 4.4e-04 | Click! |
Activity profile of Plag1 motif
Sorted Z-values of Plag1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_198232344 | 40.83 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr10_+_57278307 | 38.04 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr9_+_81821346 | 36.53 |
ENSRNOT00000022234
|
Plcd4
|
phospholipase C, delta 4 |
| chr1_+_198655742 | 35.05 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_-_220136470 | 34.75 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr1_-_215834704 | 31.82 |
ENSRNOT00000073850
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_88066101 | 31.17 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr1_+_101427195 | 29.85 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chrX_-_78496847 | 28.28 |
ENSRNOT00000003265
|
Itm2a
|
integral membrane protein 2A |
| chr5_+_148528725 | 26.86 |
ENSRNOT00000017325
|
Fabp3
|
fatty acid binding protein 3 |
| chr10_-_45480999 | 26.83 |
ENSRNOT00000078353
ENSRNOT00000084697 ENSRNOT00000087926 |
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr19_+_880024 | 26.64 |
ENSRNOT00000061648
|
Cmtm2a
|
CKLF-like MARVEL transmembrane domain containing 2A |
| chr7_-_50638798 | 26.62 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr18_+_36371041 | 26.02 |
ENSRNOT00000025408
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr10_+_14094754 | 25.25 |
ENSRNOT00000019660
|
Rpl3l
|
ribosomal protein L3-like |
| chr1_-_222246765 | 24.49 |
ENSRNOT00000087285
ENSRNOT00000028731 |
Dnajc4
|
DnaJ heat shock protein family (Hsp40) member C4 |
| chr4_+_56674832 | 24.14 |
ENSRNOT00000060309
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr2_+_188844073 | 24.11 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr6_-_99783047 | 24.04 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr13_+_51034256 | 23.47 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr1_+_81372650 | 23.36 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr17_-_71105286 | 22.97 |
ENSRNOT00000025901
|
Prkcq
|
protein kinase C, theta |
| chrX_+_159112880 | 22.68 |
ENSRNOT00000084594
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_148193710 | 22.33 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chrX_-_72133692 | 21.47 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr1_+_72889270 | 21.40 |
ENSRNOT00000058843
ENSRNOT00000034957 |
Tnnt1
|
troponin T1, slow skeletal type |
| chr1_+_197839430 | 21.05 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr13_-_52197205 | 20.46 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr13_+_104284660 | 20.17 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr18_-_28454756 | 20.15 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr1_-_73753128 | 20.13 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr8_+_48422036 | 19.84 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr1_-_205030567 | 19.81 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
| chr9_+_14529218 | 19.66 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr9_+_99998275 | 19.21 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr14_-_77810147 | 19.14 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr20_+_13778178 | 18.34 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr19_-_43290363 | 17.92 |
ENSRNOT00000024248
|
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chrX_-_29648359 | 17.90 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr10_-_82209459 | 17.68 |
ENSRNOT00000004377
|
Spata20
|
spermatogenesis associated 20 |
| chr3_+_148386189 | 17.62 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chrX_+_114929029 | 17.49 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr2_+_198797159 | 16.93 |
ENSRNOT00000056225
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr1_+_84328114 | 16.90 |
ENSRNOT00000028266
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr2_-_210241455 | 16.88 |
ENSRNOT00000086420
|
Kcnc4
|
potassium voltage-gated channel subfamily C member 4 |
| chr1_+_215666628 | 16.28 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr1_+_101324652 | 16.27 |
ENSRNOT00000028116
|
Hrc
|
histidine rich calcium binding protein |
| chr6_-_133716847 | 16.00 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr8_+_56179816 | 15.94 |
ENSRNOT00000059078
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr16_-_20686317 | 15.76 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr6_-_135412312 | 15.28 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr5_+_167109691 | 15.23 |
ENSRNOT00000055536
|
Gpr157
|
G protein-coupled receptor 157 |
| chr7_-_139483997 | 15.18 |
ENSRNOT00000086062
|
Col2a1
|
collagen type II alpha 1 chain |
| chr19_-_37427989 | 15.15 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_-_55673704 | 15.14 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr10_-_58973020 | 15.04 |
ENSRNOT00000020379
|
Smtnl2
|
smoothelin-like 2 |
| chr1_+_222519615 | 15.00 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr1_+_36185916 | 14.89 |
ENSRNOT00000041762
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr7_+_15785410 | 14.83 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr9_+_82556573 | 14.66 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr10_-_95934345 | 14.62 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr1_-_84491466 | 14.39 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr7_-_70556827 | 14.38 |
ENSRNOT00000007721
|
Kif5a
|
kinesin family member 5A |
| chr10_+_56546710 | 14.21 |
ENSRNOT00000023003
|
Ybx2
|
Y box binding protein 2 |
| chr6_+_105364668 | 14.11 |
ENSRNOT00000009513
ENSRNOT00000087090 |
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr14_+_60764409 | 14.09 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr13_-_91872954 | 14.05 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr18_-_76878981 | 14.01 |
ENSRNOT00000088054
|
Kcng2
|
potassium voltage-gated channel modifier subfamily G member 2 |
| chr17_-_19580929 | 13.90 |
ENSRNOT00000023613
|
Gmpr
|
guanosine monophosphate reductase |
| chr1_-_101351879 | 13.86 |
ENSRNOT00000028142
|
Ppfia3
|
PTPRF interacting protein alpha 3 |
| chr7_-_70552897 | 13.65 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr15_+_42960307 | 13.53 |
ENSRNOT00000012528
|
Trim35
|
tripartite motif-containing 35 |
| chr15_-_42638392 | 13.49 |
ENSRNOT00000022020
|
Scara3
|
scavenger receptor class A, member 3 |
| chr10_+_94170766 | 13.48 |
ENSRNOT00000010627
|
Ace
|
angiotensin I converting enzyme |
| chr20_+_6923489 | 13.39 |
ENSRNOT00000093373
ENSRNOT00000000632 |
Pi16
|
peptidase inhibitor 16 |
| chr5_-_59025631 | 13.37 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr15_-_104168564 | 13.34 |
ENSRNOT00000093385
ENSRNOT00000038596 |
Dzip1
|
DAZ interacting zinc finger protein 1 |
| chr8_+_118013612 | 13.17 |
ENSRNOT00000056166
|
Map4
|
microtubule-associated protein 4 |
| chr8_+_59344083 | 13.16 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chr20_+_5049496 | 12.97 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_11383116 | 12.96 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr8_-_72841496 | 12.87 |
ENSRNOT00000057641
ENSRNOT00000040808 ENSRNOT00000085894 ENSRNOT00000024575 ENSRNOT00000048044 ENSRNOT00000024493 |
Tpm1
|
tropomyosin 1, alpha |
| chr1_+_274391932 | 12.75 |
ENSRNOT00000054685
|
Rbm20
|
RNA binding motif protein 20 |
| chr5_-_146446227 | 12.56 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr7_-_117722703 | 12.24 |
ENSRNOT00000093083
ENSRNOT00000092789 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr17_+_70684134 | 12.24 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chrX_+_39711201 | 12.17 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr20_+_7136007 | 12.10 |
ENSRNOT00000000580
|
Hmga1
|
high mobility group AT-hook 1 |
| chr3_-_113312127 | 11.95 |
ENSRNOT00000065285
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr1_+_100832324 | 11.82 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr10_-_91302155 | 11.80 |
ENSRNOT00000004371
|
Spata32
|
spermatogenesis associated 32 |
| chr1_-_128695796 | 11.74 |
ENSRNOT00000076329
|
Synm
|
synemin |
| chr4_-_133951264 | 11.71 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr19_-_10596851 | 11.53 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr14_-_84170835 | 11.52 |
ENSRNOT00000005646
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr15_-_52116001 | 11.51 |
ENSRNOT00000013284
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr2_-_187372652 | 11.49 |
ENSRNOT00000025496
ENSRNOT00000083116 |
Bcan
|
brevican |
| chr1_+_81260548 | 11.47 |
ENSRNOT00000026669
|
Smg9
|
SMG9 nonsense mediated mRNA decay factor |
| chr7_-_74901997 | 11.47 |
ENSRNOT00000039378
|
Rgs22
|
regulator of G-protein signaling 22 |
| chr5_-_25584278 | 11.44 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_-_21760292 | 11.43 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr12_-_40590361 | 11.40 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr1_+_264796812 | 11.35 |
ENSRNOT00000021171
|
Sfxn3
|
sideroflexin 3 |
| chr10_-_39405311 | 11.31 |
ENSRNOT00000074616
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr1_+_101474334 | 11.28 |
ENSRNOT00000064245
|
Tulp2
|
tubby-like protein 2 |
| chr9_-_44237117 | 11.24 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr8_+_22189600 | 11.11 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr1_-_64405149 | 11.08 |
ENSRNOT00000089944
|
Cacng7
|
calcium voltage-gated channel auxiliary subunit gamma 7 |
| chr9_+_20213588 | 11.05 |
ENSRNOT00000089341
|
LOC100911515
|
triosephosphate isomerase-like |
| chr3_-_111994337 | 11.01 |
ENSRNOT00000030502
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr3_+_48626038 | 10.99 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr7_+_140383397 | 10.96 |
ENSRNOT00000090760
|
Ccdc65
|
coiled-coil domain containing 65 |
| chr6_-_92760018 | 10.93 |
ENSRNOT00000009560
|
Trim9
|
tripartite motif-containing 9 |
| chr1_+_89491654 | 10.90 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr8_-_130224444 | 10.85 |
ENSRNOT00000026173
|
Lyzl4
|
lysozyme-like 4 |
| chr15_+_1054937 | 10.84 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr7_+_18668692 | 10.70 |
ENSRNOT00000009532
|
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr17_+_83221827 | 10.67 |
ENSRNOT00000000155
|
Plxdc2
|
plexin domain containing 2 |
| chr1_+_40816107 | 10.58 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr4_+_123801174 | 10.56 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr10_+_13061170 | 10.55 |
ENSRNOT00000004936
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chrX_-_123788898 | 10.54 |
ENSRNOT00000009123
|
Akap14
|
A-kinase anchoring protein 14 |
| chr5_+_135029955 | 10.53 |
ENSRNOT00000074860
|
LOC100911669
|
uncharacterized LOC100911669 |
| chr6_-_1942972 | 10.51 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chr8_-_109621408 | 10.50 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_-_246737997 | 10.45 |
ENSRNOT00000021719
|
Pdha2
|
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr13_-_81214494 | 10.37 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr2_-_84531192 | 10.32 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr8_-_53901358 | 10.32 |
ENSRNOT00000047666
ENSRNOT00000042281 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr1_-_170397191 | 10.22 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr3_-_11885311 | 10.21 |
ENSRNOT00000021189
ENSRNOT00000021178 |
Stxbp1
|
syntaxin binding protein 1 |
| chr17_+_5311274 | 10.18 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr1_+_101682172 | 10.18 |
ENSRNOT00000028540
|
Car11
|
carbonic anhydrase 11 |
| chr4_-_157679962 | 10.15 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_215846911 | 10.11 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_18491384 | 10.06 |
ENSRNOT00000079138
ENSRNOT00000014917 |
Lama2
|
laminin subunit alpha 2 |
| chr19_-_62158327 | 10.01 |
ENSRNOT00000075053
|
Ccdc7
|
coiled-coil domain containing 7 |
| chr19_+_56220755 | 9.88 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr11_+_86552022 | 9.86 |
ENSRNOT00000002597
|
Tbx1
|
T-box 1 |
| chr16_+_21050243 | 9.78 |
ENSRNOT00000064308
|
Ncan
|
neurocan |
| chr5_-_58078545 | 9.75 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr5_-_57372239 | 9.72 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr3_+_65672058 | 9.72 |
ENSRNOT00000057901
|
RGD1564319
|
similar to TF-1 apoptosis related protein 19 |
| chr1_-_81534140 | 9.69 |
ENSRNOT00000027202
|
Tex101
|
testis expressed 101 |
| chr1_-_84254645 | 9.68 |
ENSRNOT00000077651
|
Sptbn4
|
spectrin, beta, non-erythrocytic 4 |
| chr5_+_122390522 | 9.64 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chrX_-_38196060 | 9.63 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr2_-_62634785 | 9.57 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr15_-_14737704 | 9.47 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr5_-_139227196 | 9.42 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr4_-_157798868 | 9.35 |
ENSRNOT00000044425
|
Tuba3b
|
tubulin, alpha 3B |
| chrX_-_82986051 | 9.26 |
ENSRNOT00000077587
|
Hdx
|
highly divergent homeobox |
| chr2_-_45518502 | 9.22 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr12_+_23151180 | 9.20 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr3_-_148932878 | 9.09 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr2_+_188528979 | 9.05 |
ENSRNOT00000087934
|
Thbs3
|
thrombospondin 3 |
| chr6_-_33738825 | 9.05 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chrX_+_45965301 | 9.01 |
ENSRNOT00000005141
|
Fam47a
|
family with sequence similarity 47, member A |
| chr10_+_90134193 | 9.00 |
ENSRNOT00000028375
|
G6pc3
|
glucose 6 phosphatase catalytic subunit 3 |
| chr19_-_37370429 | 9.00 |
ENSRNOT00000022497
|
Kctd19
|
potassium channel tetramerization domain containing 19 |
| chr19_-_10360310 | 8.95 |
ENSRNOT00000087100
|
Katnb1
|
katanin regulatory subunit B1 |
| chr10_-_74679858 | 8.84 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr2_-_189765415 | 8.72 |
ENSRNOT00000020815
|
Slc27a3
|
solute carrier family 27 member 3 |
| chr7_-_140483693 | 8.71 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr5_+_61565064 | 8.65 |
ENSRNOT00000090186
|
Tdrd7
|
tudor domain containing 7 |
| chr4_+_7377563 | 8.60 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr1_-_216663720 | 8.53 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr3_+_6773813 | 8.47 |
ENSRNOT00000065953
ENSRNOT00000013443 |
Olfm1
|
olfactomedin 1 |
| chr20_-_5533600 | 8.43 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr1_+_102915191 | 8.43 |
ENSRNOT00000017851
|
Ldhc
|
lactate dehydrogenase C |
| chr10_-_85938085 | 8.31 |
ENSRNOT00000043561
|
Plxdc1
|
plexin domain containing 1 |
| chr5_-_151709877 | 8.29 |
ENSRNOT00000080602
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr9_+_20213776 | 8.24 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr7_-_72328128 | 8.18 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr3_-_79743737 | 8.14 |
ENSRNOT00000013584
|
Ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr7_-_11777503 | 8.10 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chrX_-_139329975 | 8.06 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chrX_-_130794673 | 8.03 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr18_-_86878142 | 7.97 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chr1_+_165237847 | 7.87 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chrX_+_69730242 | 7.86 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr10_-_89084885 | 7.77 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr5_+_108278217 | 7.77 |
ENSRNOT00000038972
|
Dmrta1
|
DMRT-like family A1 |
| chr5_-_73494030 | 7.76 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr5_-_22769907 | 7.72 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr7_-_31824064 | 7.67 |
ENSRNOT00000011494
ENSRNOT00000080824 |
Slc25a3
|
solute carrier family 25 member 3 |
| chr7_-_115045802 | 7.67 |
ENSRNOT00000040236
ENSRNOT00000071818 |
Mroh5
|
maestro heat-like repeat family member 5 |
| chr13_+_48607308 | 7.65 |
ENSRNOT00000063882
|
Slc41a1
|
solute carrier family 41 member 1 |
| chr1_+_101783621 | 7.64 |
ENSRNOT00000067679
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr6_-_26629654 | 7.59 |
ENSRNOT00000031244
|
Dnajc5g
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chr5_+_58937615 | 7.57 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr8_-_49045154 | 7.56 |
ENSRNOT00000088034
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr15_+_33600102 | 7.55 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr20_+_3555135 | 7.54 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_101883744 | 7.49 |
ENSRNOT00000028635
|
Syngr4
|
synaptogyrin 4 |
| chr5_+_61564864 | 7.46 |
ENSRNOT00000085738
|
Tdrd7
|
tudor domain containing 7 |
| chr1_-_129217058 | 7.41 |
ENSRNOT00000086439
|
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr9_+_47987 | 7.38 |
ENSRNOT00000066582
|
Efhb
|
EF hand domain family, member B |
| chr7_+_128500011 | 7.35 |
ENSRNOT00000074625
|
Fam19a5
|
family with sequence similarity 19 member A5, C-C motif chemokine like |
| chr5_-_164971903 | 7.33 |
ENSRNOT00000067059
|
Fbxo44
|
F-box protein 44 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plag1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.5 | 52.4 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 9.3 | 28.0 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 9.0 | 26.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 7.1 | 21.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 7.0 | 21.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 6.7 | 26.6 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 6.4 | 19.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 6.0 | 41.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 5.7 | 23.0 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 5.7 | 28.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 5.6 | 16.9 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 5.4 | 16.3 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 5.0 | 20.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 4.9 | 19.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 4.9 | 19.6 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 4.5 | 31.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 4.3 | 21.5 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) |
| 4.3 | 12.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 3.9 | 11.7 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 3.6 | 17.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 3.5 | 10.5 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 3.4 | 10.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 3.4 | 10.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.4 | 13.5 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 3.3 | 19.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 3.3 | 61.8 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 3.2 | 15.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 3.0 | 15.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 3.0 | 12.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 3.0 | 9.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.9 | 11.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 2.9 | 8.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 2.8 | 8.4 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 2.8 | 16.6 | GO:0071306 | cellular response to vitamin E(GO:0071306) |
| 2.7 | 8.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 2.5 | 4.9 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 2.4 | 7.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 2.4 | 7.3 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 2.4 | 14.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 2.3 | 23.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 2.3 | 9.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 2.3 | 6.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 2.3 | 11.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) |
| 2.3 | 6.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.2 | 6.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 2.2 | 31.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 2.2 | 4.4 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 2.2 | 6.5 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 2.1 | 6.4 | GO:0048749 | compound eye development(GO:0048749) |
| 2.1 | 18.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.1 | 8.2 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 2.0 | 6.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 2.0 | 9.8 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 1.9 | 9.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.8 | 5.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.8 | 7.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 1.8 | 5.5 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.8 | 5.4 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 1.8 | 7.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.7 | 6.7 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 1.7 | 13.4 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 1.6 | 9.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.5 | 7.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.5 | 9.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.5 | 4.4 | GO:0018201 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 1.4 | 2.9 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.4 | 5.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.4 | 8.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.4 | 20.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.3 | 2.7 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 1.3 | 2.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.3 | 5.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.3 | 6.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.3 | 7.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.3 | 5.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.3 | 6.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 1.2 | 13.6 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 1.2 | 4.8 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.2 | 3.6 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.2 | 13.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.2 | 15.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 1.2 | 19.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 1.2 | 4.7 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.2 | 8.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.2 | 18.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.2 | 3.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.1 | 11.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.1 | 19.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 1.1 | 3.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.1 | 2.2 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 1.1 | 3.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 1.1 | 10.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 1.1 | 3.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 1.1 | 10.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.0 | 3.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.0 | 14.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.0 | 7.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.0 | 6.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.0 | 4.0 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 1.0 | 4.0 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.0 | 9.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 1.0 | 5.9 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.0 | 4.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.0 | 2.9 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.9 | 5.7 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.9 | 5.5 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.9 | 17.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.9 | 10.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.9 | 12.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.9 | 8.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.9 | 3.4 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.9 | 34.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.9 | 3.4 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.9 | 5.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.8 | 2.4 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.8 | 5.7 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.8 | 1.6 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.8 | 1.6 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.8 | 3.9 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.8 | 3.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.8 | 5.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.8 | 7.8 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.8 | 5.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.8 | 7.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 12.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.7 | 20.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.7 | 5.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.7 | 2.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.7 | 30.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.7 | 5.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.7 | 11.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.7 | 4.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.7 | 13.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.7 | 4.7 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.7 | 6.0 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.7 | 29.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.7 | 2.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) cell differentiation involved in salivary gland development(GO:0060689) |
| 0.6 | 7.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 14.6 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.6 | 7.6 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.6 | 11.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.6 | 3.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.6 | 14.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.6 | 5.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 4.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.6 | 3.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.6 | 6.9 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.6 | 1.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.6 | 11.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.6 | 1.7 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.5 | 4.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 4.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 22.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.5 | 13.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 11.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.5 | 11.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.5 | 42.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 5.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.5 | 5.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.5 | 7.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.5 | 9.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.5 | 16.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 17.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.5 | 1.0 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.5 | 1.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.5 | 0.9 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.5 | 6.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 4.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.4 | 0.9 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.4 | 12.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.4 | 5.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.4 | 1.3 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.4 | 3.0 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.4 | 2.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.4 | 9.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 4.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.4 | 4.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 2.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 2.8 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 5.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 1.9 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 5.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 4.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.4 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 3.0 | GO:0007512 | adult heart development(GO:0007512) |
| 0.4 | 27.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 7.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.3 | 1.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 5.2 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.3 | 0.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.3 | 6.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 7.0 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 8.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.3 | 3.3 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.3 | 4.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 6.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.3 | 12.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.3 | 13.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.3 | 6.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 2.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 0.9 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.3 | 3.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.9 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.3 | 16.1 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.3 | 6.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.3 | 8.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.3 | 7.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.3 | 6.7 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.3 | 0.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 10.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.3 | 1.7 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.3 | 1.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 3.7 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.3 | 2.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.3 | 13.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.3 | 8.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.3 | 7.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 2.7 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.3 | 0.8 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 6.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.3 | 4.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 4.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 15.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.3 | 9.4 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.3 | 3.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.3 | 10.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.3 | 9.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.3 | 6.9 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.3 | 33.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 2.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 7.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.2 | 1.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 2.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 10.7 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.2 | 2.6 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.2 | 19.3 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 3.9 | GO:0032402 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.2 | 11.1 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.2 | 3.2 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.2 | 5.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 3.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.2 | 3.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 8.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 0.4 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.2 | 4.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.2 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 5.6 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.2 | 1.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.2 | 1.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 1.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 2.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.9 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 1.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) regulation of male germ cell proliferation(GO:2000254) |
| 0.1 | 5.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 5.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 17.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 2.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 1.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 6.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 4.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.5 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 4.0 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 5.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.9 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 2.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 9.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.7 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 2.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.6 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 2.9 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 3.4 | GO:0001508 | action potential(GO:0001508) |
| 0.1 | 4.6 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 2.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 4.5 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.1 | 0.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 2.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 1.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 1.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 1.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 1.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.6 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 5.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 14.6 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 1.9 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 2.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 3.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 7.3 | GO:0050803 | regulation of synapse structure or activity(GO:0050803) |
| 0.1 | 0.8 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 7.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 5.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 3.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.6 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 38.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 5.3 | 15.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 4.9 | 14.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.6 | 13.9 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 4.4 | 26.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 4.0 | 20.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 3.8 | 11.5 | GO:1990923 | PET complex(GO:1990923) |
| 3.7 | 26.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 3.5 | 31.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 3.4 | 33.7 | GO:0008091 | spectrin(GO:0008091) |
| 3.3 | 96.3 | GO:0031430 | M band(GO:0031430) |
| 3.3 | 9.8 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 3.1 | 37.7 | GO:0005861 | troponin complex(GO:0005861) |
| 2.6 | 10.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 2.4 | 7.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.3 | 11.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 2.2 | 11.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.1 | 8.3 | GO:0072487 | MSL complex(GO:0072487) |
| 2.0 | 28.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.8 | 7.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.5 | 6.0 | GO:0032021 | NELF complex(GO:0032021) |
| 1.5 | 5.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.4 | 11.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.4 | 7.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.4 | 5.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.3 | 20.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.3 | 10.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.2 | 4.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.2 | 11.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.1 | 3.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.1 | 31.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 1.1 | 5.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.1 | 81.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 1.1 | 5.5 | GO:0070695 | FHF complex(GO:0070695) |
| 1.1 | 14.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.0 | 19.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 1.0 | 10.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.0 | 4.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.9 | 11.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 6.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.9 | 7.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.9 | 22.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.9 | 7.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.9 | 22.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.8 | 2.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.8 | 8.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 16.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.8 | 5.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.8 | 10.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 3.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.7 | 2.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.7 | 10.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.7 | 5.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 4.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 1.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 1.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.6 | 19.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 2.4 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.6 | 4.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.6 | 1.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.6 | 21.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 12.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.6 | 9.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.5 | 1.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 6.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 2.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.5 | 7.9 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.5 | 3.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.5 | 7.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 4.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.5 | 10.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 17.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 9.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 12.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 6.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 10.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 3.7 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 7.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 10.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.4 | 1.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 39.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 30.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 1.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 3.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 34.9 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.3 | 3.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 8.1 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 31.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 6.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 2.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 5.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 4.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 3.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 17.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 2.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 18.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 12.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 1.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.3 | 24.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 3.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 6.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 25.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 4.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 2.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 5.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 30.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 5.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 6.9 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 4.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 4.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 6.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 7.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 3.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 5.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.2 | 3.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 4.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 7.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 57.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 3.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 9.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 20.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 3.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 5.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 3.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 7.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 6.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 23.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 7.1 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 1.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0019013 | viral nucleocapsid(GO:0019013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 31.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 6.7 | 20.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 6.4 | 19.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 6.3 | 38.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 5.8 | 40.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 5.4 | 37.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 5.3 | 26.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.9 | 9.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 4.6 | 13.9 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 4.5 | 13.5 | GO:0070573 | bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 4.5 | 26.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) oleic acid binding(GO:0070538) |
| 4.4 | 17.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.3 | 13.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 4.3 | 29.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 4.0 | 12.0 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 3.9 | 11.8 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 3.8 | 19.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 3.8 | 11.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 3.6 | 14.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 3.3 | 19.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 3.2 | 9.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 3.0 | 11.9 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 2.9 | 20.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.9 | 11.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 2.7 | 27.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 2.7 | 26.8 | GO:0031432 | titin binding(GO:0031432) |
| 2.6 | 15.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 2.5 | 10.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.5 | 15.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.4 | 7.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 2.3 | 9.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 2.3 | 11.4 | GO:0015142 | tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 2.2 | 102.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 2.2 | 6.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 2.1 | 47.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 2.1 | 8.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.0 | 43.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.9 | 20.9 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.9 | 5.6 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 1.8 | 5.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.8 | 5.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 1.8 | 14.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 1.8 | 9.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.8 | 12.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.8 | 28.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.7 | 6.9 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.7 | 5.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.6 | 6.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.5 | 10.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.5 | 17.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.5 | 4.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.4 | 6.8 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 1.3 | 25.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 1.3 | 5.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.2 | 9.9 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 1.2 | 3.6 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.2 | 4.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.1 | 5.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.1 | 4.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.1 | 32.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.1 | 7.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.1 | 6.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.1 | 17.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.0 | 6.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.0 | 4.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 1.0 | 9.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.0 | 5.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.0 | 13.2 | GO:0019841 | retinol binding(GO:0019841) |
| 1.0 | 5.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 8.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 1.0 | 2.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.0 | 24.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.0 | 2.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.9 | 2.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.9 | 2.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.9 | 5.3 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.8 | 10.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 21.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.8 | 3.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.7 | 3.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.7 | 32.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.7 | 10.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 5.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.7 | 14.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.7 | 6.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 8.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 7.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.7 | 6.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.6 | 6.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.6 | 2.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.6 | 2.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.6 | 11.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.6 | 6.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 18.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 82.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.5 | 5.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 11.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 4.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.5 | 12.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.5 | 3.5 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.5 | 7.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 3.4 | GO:0050700 | LIM domain binding(GO:0030274) CARD domain binding(GO:0050700) |
| 0.5 | 1.4 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.5 | 1.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 12.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 7.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 4.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 3.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 1.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.4 | 32.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.4 | 14.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.4 | 6.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 5.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 4.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 10.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 4.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.4 | 8.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 2.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 3.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.4 | 10.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 4.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 21.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 3.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.3 | 7.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.3 | 8.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 2.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 85.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 1.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 3.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 6.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 8.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 10.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.3 | 4.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 43.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 5.8 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 6.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 7.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 15.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 4.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 7.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 0.4 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 8.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 49.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 4.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 7.7 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.1 | 2.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.0 | GO:0015491 | cation:cation antiporter activity(GO:0015491) |
| 0.1 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 5.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 11.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 9.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 8.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 3.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 4.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 4.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 3.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 19.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 18.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0019199 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) transmembrane receptor protein kinase activity(GO:0019199) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0099516 | ion antiporter activity(GO:0099516) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.9 | 27.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.9 | 31.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.9 | 34.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.8 | 14.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.7 | 13.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 18.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.7 | 43.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.7 | 8.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.7 | 24.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 50.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.7 | 10.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 16.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 24.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.5 | 10.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 26.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 15.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 14.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 19.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 6.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 8.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.4 | 23.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 3.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 2.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 4.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 19.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 10.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 17.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 4.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 2.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 6.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 9.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 3.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 29.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 6.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 5.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 6.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 3.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 3.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 6.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 4.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 41.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 2.8 | 110.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.7 | 8.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.6 | 62.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.5 | 21.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.2 | 13.9 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 1.1 | 39.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.1 | 19.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 1.0 | 29.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 1.0 | 50.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.9 | 28.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 13.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 12.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.8 | 9.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 12.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 27.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.7 | 10.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.6 | 9.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.6 | 7.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 33.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.5 | 6.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.5 | 7.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 6.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 8.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 2.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.4 | 9.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 11.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.4 | 17.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 4.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 7.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 5.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 13.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 3.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 4.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 6.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 1.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 2.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 6.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 1.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 3.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 5.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 5.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 9.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 0.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 1.9 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 17.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 7.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 8.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |