Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
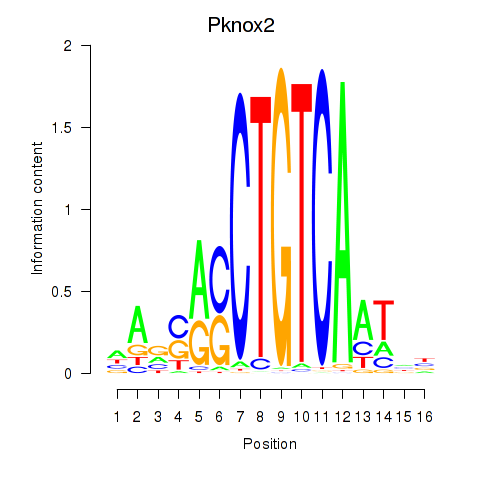
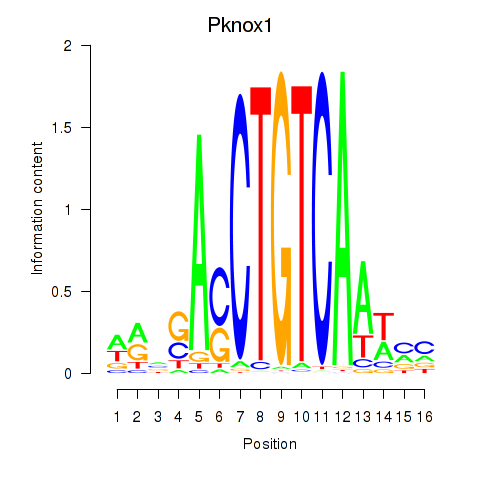
Results for Pknox2_Pknox1
Z-value: 0.76
Transcription factors associated with Pknox2_Pknox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pknox2
|
ENSRNOG00000028856 | PBX/knotted 1 homeobox 2 |
|
Pknox1
|
ENSRNOG00000001184 | PBX/knotted 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pknox1 | rn6_v1_chr20_+_10334308_10334308 | 0.41 | 1.0e-14 | Click! |
| Pknox2 | rn6_v1_chr8_-_39551700_39551700 | -0.07 | 2.1e-01 | Click! |
Activity profile of Pknox2_Pknox1 motif
Sorted Z-values of Pknox2_Pknox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_73958480 | 52.35 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr13_+_52889737 | 46.66 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr10_+_4945911 | 30.25 |
ENSRNOT00000003420
|
Prm1
|
protamine 1 |
| chr17_-_43614844 | 29.51 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr10_-_85974644 | 18.24 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr2_-_140618405 | 15.86 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr8_-_33017854 | 15.35 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr3_+_148386189 | 15.12 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr4_+_69138525 | 14.82 |
ENSRNOT00000073589
|
Trbv1
|
T cell receptor beta, variable 1 |
| chr9_+_14529218 | 14.49 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr7_+_66595742 | 12.97 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr20_+_46667121 | 10.04 |
ENSRNOT00000089611
|
Sesn1
|
sestrin 1 |
| chr20_+_46667454 | 9.72 |
ENSRNOT00000065890
|
Sesn1
|
sestrin 1 |
| chr15_+_34251606 | 9.57 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr12_-_17904254 | 9.41 |
ENSRNOT00000060192
|
LOC680273
|
similar to Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead-related transcription factor 7) (FREAC-7) |
| chr4_-_62526724 | 8.47 |
ENSRNOT00000038950
|
RGD1565367
|
similar to Solute carrier family 23, member 2 (Sodium-dependent vitamin C transporter 2) |
| chr16_+_72086878 | 8.39 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chr1_+_264893162 | 8.10 |
ENSRNOT00000021714
|
Tlx1
|
T-cell leukemia, homeobox 1 |
| chr4_+_127164453 | 7.71 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr6_-_103605256 | 7.41 |
ENSRNOT00000006005
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr2_-_105047984 | 7.09 |
ENSRNOT00000014970
|
Cpa3
|
carboxypeptidase A3 |
| chr20_-_3728844 | 6.98 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr20_-_5212624 | 6.78 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr20_+_3995544 | 6.33 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr8_-_67869019 | 6.25 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chrX_-_138435391 | 5.80 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr13_+_97838361 | 5.76 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr4_-_99125111 | 5.49 |
ENSRNOT00000009184
|
Smyd1
|
SET and MYND domain containing 1 |
| chr10_+_105498728 | 5.41 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr5_-_146446227 | 5.34 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr1_+_75693232 | 4.85 |
ENSRNOT00000020090
|
Cabp5
|
calcium binding protein 5 |
| chr9_+_81518176 | 4.68 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr5_+_126511350 | 4.49 |
ENSRNOT00000011408
|
Ssbp3
|
single stranded DNA binding protein 3 |
| chr6_+_129052503 | 4.44 |
ENSRNOT00000044152
|
RGD1565462
|
similar to BC049975 protein |
| chr3_+_175144495 | 4.40 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr7_-_120967490 | 4.40 |
ENSRNOT00000046399
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr3_-_125213607 | 4.34 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr7_-_27136652 | 4.30 |
ENSRNOT00000086905
|
Hcfc2
|
host cell factor C2 |
| chr11_+_80358211 | 4.06 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr7_-_60743328 | 3.98 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr9_-_45206427 | 3.84 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr8_-_76239995 | 3.75 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr8_+_94122728 | 3.66 |
ENSRNOT00000014783
|
Dopey1
|
dopey family member 1 |
| chr11_+_84745904 | 3.65 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr9_-_94201084 | 3.65 |
ENSRNOT00000026190
|
Alpi
|
alkaline phosphatase, intestinal |
| chr9_+_94228960 | 3.30 |
ENSRNOT00000083137
ENSRNOT00000080897 |
Akp3
|
alkaline phosphatase 3, intestine, not Mn requiring |
| chr10_-_87928251 | 3.19 |
ENSRNOT00000064120
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr1_-_201981229 | 3.17 |
ENSRNOT00000077246
ENSRNOT00000035531 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr6_+_42947334 | 3.16 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr15_-_88670349 | 3.16 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr9_+_17163354 | 2.97 |
ENSRNOT00000026049
|
Polh
|
DNA polymerase eta |
| chr10_+_105393072 | 2.95 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr14_+_83412968 | 2.84 |
ENSRNOT00000024993
|
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr20_+_31035729 | 2.81 |
ENSRNOT00000000672
|
Nodal
|
nodal growth differentiation factor |
| chr10_-_46969406 | 2.52 |
ENSRNOT00000006753
|
Flii
|
FLII, actin remodeling protein |
| chr3_-_13978224 | 2.52 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr4_-_80313411 | 2.48 |
ENSRNOT00000041621
|
RGD1565415
|
similar to ribosomal protein L27a |
| chr15_-_32925673 | 2.47 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr18_+_44468784 | 2.40 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chrX_+_136460215 | 2.33 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr11_-_82366505 | 2.29 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr2_+_140471690 | 2.28 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr10_+_66099531 | 2.27 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr10_+_90731148 | 2.25 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr10_+_90731865 | 2.20 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr12_+_660011 | 2.17 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr2_-_43999107 | 2.14 |
ENSRNOT00000065473
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr10_+_88245532 | 2.13 |
ENSRNOT00000019863
|
Gast
|
gastrin |
| chr17_-_10208360 | 1.99 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr10_-_97647822 | 1.97 |
ENSRNOT00000065781
|
AC106648.1
|
|
| chr4_+_178617401 | 1.95 |
ENSRNOT00000042619
|
AABR07062466.1
|
|
| chr1_-_80073223 | 1.89 |
ENSRNOT00000021456
|
Gipr
|
gastric inhibitory polypeptide receptor |
| chr14_+_5928737 | 1.87 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr17_+_6786348 | 1.83 |
ENSRNOT00000083408
|
Gkap1
|
G kinase anchoring protein 1 |
| chr1_+_15180328 | 1.81 |
ENSRNOT00000050656
|
Il20ra
|
interleukin 20 receptor subunit alpha |
| chr4_+_129574264 | 1.79 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr6_+_52265930 | 1.60 |
ENSRNOT00000087513
ENSRNOT00000067849 |
Sypl1
|
synaptophysin-like 1 |
| chr5_-_77749613 | 1.55 |
ENSRNOT00000075988
|
Mup5
|
major urinary protein 5 |
| chr14_+_42401706 | 1.53 |
ENSRNOT00000090148
|
Bend4
|
BEN domain containing 4 |
| chr13_-_95250235 | 1.51 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr6_+_132090218 | 1.47 |
ENSRNOT00000083208
|
Ccnk
|
cyclin K |
| chr1_+_93242050 | 1.46 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr2_+_198797159 | 1.45 |
ENSRNOT00000056225
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr2_-_188727670 | 1.42 |
ENSRNOT00000074920
|
Lenep
|
lens epithelial protein |
| chr20_+_46428124 | 1.41 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr7_+_12146642 | 1.40 |
ENSRNOT00000085899
|
Tcf3
|
transcription factor 3 |
| chr2_+_155555840 | 1.38 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr3_-_23297774 | 1.33 |
ENSRNOT00000019162
|
Rpl35
|
ribosomal protein L35 |
| chr6_+_132090633 | 1.32 |
ENSRNOT00000057171
|
Ccnk
|
cyclin K |
| chr6_-_137733026 | 1.25 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr10_+_84214358 | 1.21 |
ENSRNOT00000011416
|
Hoxb1
|
homeo box B1 |
| chr3_-_8979889 | 1.20 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chrX_+_18998532 | 1.20 |
ENSRNOT00000004213
|
AABR07037343.1
|
|
| chr18_-_24312950 | 1.18 |
ENSRNOT00000051911
|
LOC686074
|
similar to 60S ribosomal protein L35 |
| chr3_+_65816569 | 1.17 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr15_+_51222954 | 1.17 |
ENSRNOT00000064428
ENSRNOT00000088928 |
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr14_-_105055421 | 1.15 |
ENSRNOT00000008274
|
Lgalsl
|
galectin-like |
| chr10_+_87774552 | 1.12 |
ENSRNOT00000044342
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr7_-_105592804 | 1.09 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr5_+_155649217 | 1.08 |
ENSRNOT00000018064
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr7_+_108613739 | 0.99 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chr15_-_28081465 | 0.98 |
ENSRNOT00000033739
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr7_+_5324469 | 0.91 |
ENSRNOT00000071164
|
Olr901
|
olfactory receptor 901 |
| chr12_-_17186679 | 0.90 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr12_+_7464155 | 0.80 |
ENSRNOT00000078798
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr20_-_41209728 | 0.78 |
ENSRNOT00000000657
|
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr13_-_104156454 | 0.71 |
ENSRNOT00000031419
|
AABR07022046.1
|
|
| chr7_-_3752134 | 0.63 |
ENSRNOT00000067319
|
Olr880
|
olfactory receptor 880 |
| chr7_+_123531682 | 0.60 |
ENSRNOT00000010606
|
Wbp2nl
|
WBP2 N-terminal like |
| chr10_+_11595044 | 0.57 |
ENSRNOT00000007079
|
Crebbp
|
CREB binding protein |
| chrX_-_80142902 | 0.48 |
ENSRNOT00000071650
|
Hmgn5
|
high mobility group nucleosome binding domain 5 |
| chr14_-_81254637 | 0.46 |
ENSRNOT00000058166
|
Htt
|
huntingtin |
| chr7_-_11266630 | 0.45 |
ENSRNOT00000027926
|
Cactin
|
cactin, spliceosome C complex subunit |
| chr1_+_230030914 | 0.43 |
ENSRNOT00000075730
|
Olr354
|
olfactory receptor 354 |
| chr17_+_3385851 | 0.43 |
ENSRNOT00000061486
|
Cts8
|
cathepsin 8 |
| chr14_+_83897117 | 0.42 |
ENSRNOT00000026569
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr2_-_118989127 | 0.27 |
ENSRNOT00000014871
|
Gnb4
|
G protein subunit beta 4 |
| chr9_-_17163170 | 0.24 |
ENSRNOT00000025921
|
Xpo5
|
exportin 5 |
| chr14_-_84820415 | 0.21 |
ENSRNOT00000057501
|
Mtmr3
|
myotubularin related protein 3 |
| chr5_-_159371313 | 0.18 |
ENSRNOT00000009921
|
Padi1
|
peptidyl arginine deiminase 1 |
| chr10_+_36715565 | 0.14 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr14_-_1461543 | 0.12 |
ENSRNOT00000075656
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr1_-_198320075 | 0.06 |
ENSRNOT00000081291
|
Taok2
|
TAO kinase 2 |
| chr2_+_211337271 | 0.06 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr14_+_79261092 | 0.02 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pknox2_Pknox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 46.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 5.0 | 15.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 3.6 | 14.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 3.2 | 13.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 2.1 | 6.3 | GO:0002585 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 1.6 | 15.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 1.5 | 4.4 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.4 | 5.8 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 1.4 | 2.8 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 1.4 | 9.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.4 | 6.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.3 | 4.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) response to formaldehyde(GO:1904404) |
| 1.3 | 19.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.2 | 4.7 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 1.1 | 5.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.9 | 30.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.8 | 7.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.7 | 4.5 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.7 | 7.7 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.6 | 16.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.6 | 6.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 1.1 | GO:0030237 | female sex determination(GO:0030237) |
| 0.4 | 3.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.4 | 1.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.4 | 18.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.4 | 4.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 2.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 1.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 2.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 47.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.3 | 22.8 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.3 | 2.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 2.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 1.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.2 | 3.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.2 | 1.4 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.2 | 1.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 8.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 5.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 5.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 1.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 3.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.6 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 7.2 | GO:0048536 | spleen development(GO:0048536) |
| 0.2 | 4.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 3.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 0.5 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 3.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.9 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.1 | 0.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 2.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 4.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 4.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.8 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 2.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 3.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.8 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 7.9 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 19.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.9 | 13.1 | GO:0042825 | TAP complex(GO:0042825) |
| 1.4 | 64.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.9 | 2.8 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.9 | 29.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.8 | 4.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.8 | 52.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.6 | 4.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 1.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 31.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 2.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 7.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 2.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 7.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 12.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 6.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 6.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 6.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 15.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 16.6 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 7.2 | GO:0016604 | nuclear body(GO:0016604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 3.4 | 64.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 3.3 | 19.8 | GO:0070728 | leucine binding(GO:0070728) |
| 2.6 | 13.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 2.4 | 14.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.8 | 5.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 1.6 | 6.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.4 | 4.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.9 | 2.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.9 | 6.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.8 | 4.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.7 | 5.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.7 | 4.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.6 | 1.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 2.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 1.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 31.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.3 | 13.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.3 | 7.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 10.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 1.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 3.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 12.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 21.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 35.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 6.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 57.6 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 4.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 7.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 15.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 2.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 5.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 16.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 22.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 4.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 29.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.5 | 64.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.3 | 52.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 6.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 6.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 5.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 4.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 5.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 5.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 4.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 4.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 6.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |