Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pitx2_Otx2
Z-value: 1.22
Transcription factors associated with Pitx2_Otx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
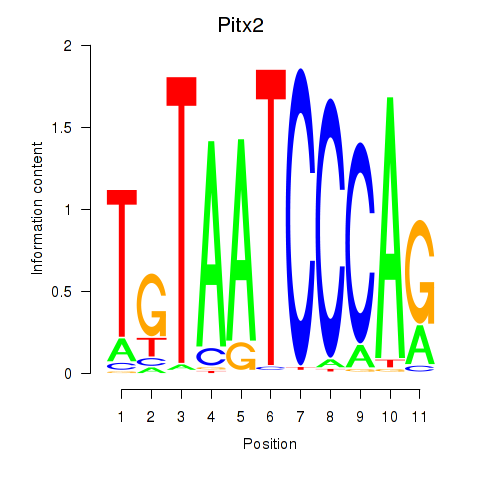
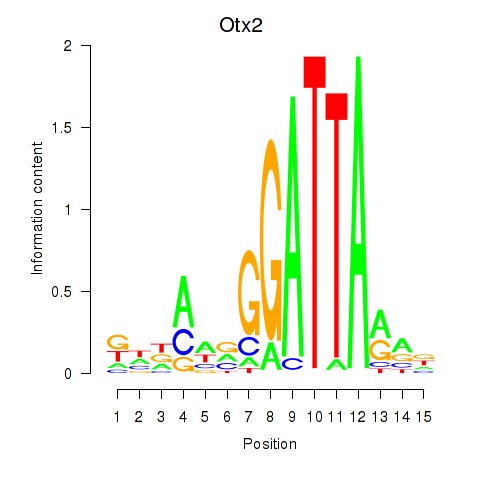
Pitx2
|
ENSRNOG00000010681 | paired-like homeodomain 2 |
|
Otx2
|
ENSRNOG00000056186 | orthodenticle homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx2 | rn6_v1_chr15_-_25521393_25521393 | 0.17 | 3.0e-03 | Click! |
| Pitx2 | rn6_v1_chr2_+_233602732_233602732 | 0.05 | 3.4e-01 | Click! |
Activity profile of Pitx2_Otx2 motif
Sorted Z-values of Pitx2_Otx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_56591715 | 42.98 |
ENSRNOT00000041207
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr3_-_3266260 | 39.65 |
ENSRNOT00000034538
|
Glt6d1
|
glycosyltransferase 6 domain containing 1 |
| chr7_-_145048283 | 38.82 |
ENSRNOT00000055275
|
Gtsf1
|
gametocyte specific factor 1 |
| chr4_-_6062641 | 37.67 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr19_+_14835822 | 37.25 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr1_-_88162583 | 30.40 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chrX_+_78991223 | 29.99 |
ENSRNOT00000031200
|
Fam46d
|
family with sequence similarity 46, member D |
| chr1_-_85317968 | 29.97 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr14_+_81858737 | 29.69 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr8_+_115213471 | 29.36 |
ENSRNOT00000017570
|
Iqcf5
|
IQ motif containing F5 |
| chr8_-_67232298 | 29.12 |
ENSRNOT00000049441
|
Spesp1
|
sperm equatorial segment protein 1 |
| chr3_+_170994038 | 28.77 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr20_+_30791422 | 28.45 |
ENSRNOT00000047394
ENSRNOT00000000683 |
Tbata
|
thymus, brain and testes associated |
| chr5_+_159612762 | 28.39 |
ENSRNOT00000012147
|
Spata21
|
spermatogenesis associated 21 |
| chr15_-_40428800 | 27.07 |
ENSRNOT00000011451
|
Atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr6_-_47848026 | 26.89 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr2_+_127549331 | 26.02 |
ENSRNOT00000093416
|
Slc25a31
|
solute carrier family 25 member 31 |
| chr7_+_27620458 | 25.17 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr5_+_128190708 | 23.85 |
ENSRNOT00000012492
|
Orc1
|
origin recognition complex, subunit 1 |
| chr8_+_22947152 | 23.78 |
ENSRNOT00000016790
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr18_+_25749098 | 23.47 |
ENSRNOT00000027771
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_+_170996193 | 23.05 |
ENSRNOT00000008959
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr14_+_8182383 | 22.31 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr9_+_10760113 | 21.72 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr16_+_68633720 | 21.58 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr1_-_167005839 | 21.57 |
ENSRNOT00000027261
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr4_+_55720010 | 21.33 |
ENSRNOT00000063987
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr14_+_33247274 | 21.15 |
ENSRNOT00000075226
|
Spink2
|
serine peptidase inhibitor, Kazal type 2 |
| chr4_-_157433467 | 20.88 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr1_+_99447561 | 20.79 |
ENSRNOT00000070899
ENSRNOT00000073798 |
Izumo2
|
IZUMO family member 2 |
| chr20_-_3401273 | 20.37 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr11_-_87977368 | 19.89 |
ENSRNOT00000002546
|
Tmem191c
|
transmembrane protein 191C |
| chr3_-_153042395 | 19.84 |
ENSRNOT00000055232
|
Ndrg3
|
NDRG family member 3 |
| chr2_+_27905535 | 19.81 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr5_-_17061361 | 19.65 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr3_+_177188044 | 19.62 |
ENSRNOT00000022073
|
Tcea2
|
transcription elongation factor A2 |
| chr8_+_115193146 | 19.34 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chr17_+_44556039 | 19.04 |
ENSRNOT00000086540
|
Prss16
|
protease, serine 16 |
| chr11_-_27971359 | 19.01 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr3_+_2438089 | 18.98 |
ENSRNOT00000013335
|
Fam166a
|
family with sequence similarity 166, member A |
| chr11_-_1437732 | 18.82 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr16_+_71242470 | 18.71 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr13_-_74005486 | 18.56 |
ENSRNOT00000090173
|
AABR07021465.1
|
|
| chr5_-_17061837 | 18.40 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr10_-_82374171 | 18.16 |
ENSRNOT00000032693
|
Eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr1_+_102924059 | 17.96 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr6_+_95205153 | 17.93 |
ENSRNOT00000007339
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr1_-_56982554 | 17.84 |
ENSRNOT00000047157
ENSRNOT00000078312 |
Tcte3
|
t-complex-associated testis expressed 3 |
| chr5_-_144345531 | 17.78 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr12_+_14021727 | 17.48 |
ENSRNOT00000060608
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr2_-_196415530 | 17.35 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr13_+_109646455 | 17.15 |
ENSRNOT00000073985
|
LOC498316
|
hypothetical LOC498316 |
| chr14_-_1505085 | 17.15 |
ENSRNOT00000090361
|
LOC102554799
|
uncharacterized LOC102554799 |
| chr6_-_26318568 | 16.99 |
ENSRNOT00000072707
|
LOC102556504
|
titin-like |
| chr10_+_55626741 | 16.94 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr20_-_5927070 | 16.81 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr20_-_5865775 | 16.64 |
ENSRNOT00000000612
ENSRNOT00000092641 |
Srpk1
|
SRSF protein kinase 1 |
| chr11_+_66316606 | 16.48 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr3_-_63211842 | 16.44 |
ENSRNOT00000008371
ENSRNOT00000050355 |
Pde11a
|
phosphodiesterase 11A |
| chr19_+_24545318 | 16.24 |
ENSRNOT00000005071
|
Clgn
|
calmegin |
| chr4_+_124005280 | 15.49 |
ENSRNOT00000014353
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_+_92573004 | 15.28 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr19_+_9895121 | 15.28 |
ENSRNOT00000033953
|
Prss54
|
protease, serine, 54 |
| chr11_-_88177448 | 15.13 |
ENSRNOT00000085475
|
Ypel1
|
yippee-like 1 |
| chr4_-_100783750 | 15.13 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr18_-_37096132 | 15.09 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr10_-_79097807 | 15.08 |
ENSRNOT00000003421
|
Kif2b
|
kinesin family member 2B |
| chr2_+_54660722 | 15.03 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr10_+_54155876 | 15.02 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr11_+_87522971 | 14.84 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr8_-_127871192 | 14.83 |
ENSRNOT00000055928
|
Slc22a14
|
solute carrier family 22, member 14 |
| chr1_-_98490315 | 14.71 |
ENSRNOT00000056490
|
Cldnd2
|
claudin domain containing 2 |
| chr4_+_123801174 | 14.71 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr12_+_24158766 | 14.66 |
ENSRNOT00000001963
|
Ccl26
|
C-C motif chemokine ligand 26 |
| chr4_-_156945126 | 14.42 |
ENSRNOT00000064853
|
RGD1307916
|
LOC362433 |
| chr20_-_44220702 | 14.24 |
ENSRNOT00000036853
|
Fam229b
|
family with sequence similarity 229, member B |
| chr9_+_24095751 | 14.15 |
ENSRNOT00000018177
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr2_+_127625683 | 14.10 |
ENSRNOT00000015474
|
Hspa4l
|
heat shock protein 4-like |
| chr10_+_65454658 | 13.94 |
ENSRNOT00000037048
|
Proca1
|
protein interacting with cyclin A1 |
| chr2_+_123790915 | 13.93 |
ENSRNOT00000023282
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr16_-_47163898 | 13.89 |
ENSRNOT00000074490
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr10_-_12469110 | 13.86 |
ENSRNOT00000060991
|
AC129054.1
|
olfactory receptor Olr1366 |
| chr1_+_277641512 | 13.70 |
ENSRNOT00000084861
ENSRNOT00000023024 |
Tdrd1
|
tudor domain containing 1 |
| chr15_-_24199341 | 13.55 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr13_-_71276497 | 13.45 |
ENSRNOT00000029773
|
Rgsl1
|
regulator of G-protein signaling like 1 |
| chr5_-_135472116 | 13.45 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr1_-_197568165 | 13.43 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr3_+_143151739 | 13.43 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chr19_-_37528011 | 13.41 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr16_+_52169450 | 13.36 |
ENSRNOT00000019474
|
Triml1
|
tripartite motif family-like 1 |
| chr5_+_151413382 | 13.34 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr18_+_63098144 | 13.31 |
ENSRNOT00000024968
|
Cidea
|
cell death-inducing DFFA-like effector a |
| chr1_+_129613497 | 13.25 |
ENSRNOT00000074410
|
LOC102551255
|
tetraspanin-7-like |
| chr9_+_10004805 | 13.25 |
ENSRNOT00000074117
|
Slc25a41
|
solute carrier family 25, member 41 |
| chrX_-_105793306 | 13.18 |
ENSRNOT00000015806
|
Nxf2
|
nuclear RNA export factor 2 |
| chr2_-_200762492 | 13.17 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr15_+_52148379 | 13.13 |
ENSRNOT00000074912
|
Phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr17_+_59697875 | 13.07 |
ENSRNOT00000077324
|
RGD1560860
|
similar to ankyrin repeat domain 26 |
| chr1_-_100205147 | 13.07 |
ENSRNOT00000046737
|
AABR07003250.1
|
|
| chr1_+_53531076 | 13.01 |
ENSRNOT00000018015
|
Tcp10b
|
t-complex protein 10b |
| chr14_+_94776233 | 12.98 |
ENSRNOT00000070839
|
AABR07016166.1
|
|
| chr20_-_6195463 | 12.96 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr15_+_47455690 | 12.92 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr4_-_174180729 | 12.89 |
ENSRNOT00000011376
|
Plcz1
|
phospholipase C, zeta 1 |
| chr1_-_89132959 | 12.87 |
ENSRNOT00000070888
|
Pmis2
|
Pmis2, sperm specific protein |
| chr5_+_58937615 | 12.85 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr17_-_78812111 | 12.75 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr16_+_12955826 | 12.64 |
ENSRNOT00000060751
|
4930596D02Rik
|
RIKEN cDNA 4930596D02 gene |
| chr14_+_33906821 | 12.62 |
ENSRNOT00000051031
|
LOC100910138
|
serine protease inhibitor Kazal-type 2-like |
| chr16_+_66230841 | 12.61 |
ENSRNOT00000050464
|
LOC689479
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr1_-_64350338 | 12.54 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr1_+_100708693 | 12.48 |
ENSRNOT00000071054
ENSRNOT00000071503 |
LOC100911976
|
izumo sperm-egg fusion protein 2-like |
| chr3_+_170475831 | 12.46 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr2_+_30666475 | 12.46 |
ENSRNOT00000076061
|
Taf9
|
TATA-box binding protein associated factor 9 |
| chrX_+_74497262 | 12.45 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr12_+_37709860 | 12.44 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr1_+_101161252 | 12.38 |
ENSRNOT00000028064
ENSRNOT00000064184 |
Slc17a7
|
solute carrier family 17 member 7 |
| chr19_-_33424955 | 12.37 |
ENSRNOT00000016558
ENSRNOT00000089569 |
Ttc29
|
tetratricopeptide repeat domain 29 |
| chr12_+_37603986 | 12.32 |
ENSRNOT00000088006
|
Sbno1
|
strawberry notch homolog 1 |
| chr1_-_279945557 | 12.29 |
ENSRNOT00000039453
|
LOC681006
|
hypothetical protein LOC681006 |
| chr4_+_179905116 | 12.26 |
ENSRNOT00000052352
|
Tuba3a
|
tubulin, alpha 3A |
| chr10_-_40489808 | 12.21 |
ENSRNOT00000037274
|
Slc36a3
|
solute carrier family 36, member 3 |
| chr10_-_86930947 | 12.18 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr9_+_14951047 | 12.16 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr1_+_205911552 | 12.13 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr19_-_57192095 | 12.11 |
ENSRNOT00000058080
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr19_+_10024947 | 12.10 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chrX_-_73562046 | 12.07 |
ENSRNOT00000080501
|
Gm14597
|
predicted gene 14597 |
| chr1_+_99532568 | 12.03 |
ENSRNOT00000077073
|
Zfp819
|
zinc finger protein 819 |
| chr7_-_74901997 | 11.98 |
ENSRNOT00000039378
|
Rgs22
|
regulator of G-protein signaling 22 |
| chr10_-_10807079 | 11.95 |
ENSRNOT00000087029
|
LOC360479
|
similar to hypothetical protein |
| chr7_-_19088972 | 11.94 |
ENSRNOT00000049266
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr3_+_143172686 | 11.94 |
ENSRNOT00000006869
|
Cst9l
|
cystatin 9-like |
| chr8_-_13909061 | 11.88 |
ENSRNOT00000078372
ENSRNOT00000083011 |
Cep295
|
centrosomal protein 295 |
| chr1_-_54854353 | 11.81 |
ENSRNOT00000072895
|
Smok2a
|
sperm motility kinase 2A |
| chr3_+_143084563 | 11.77 |
ENSRNOT00000006362
|
Cstl1
|
cystatin-like 1 |
| chr12_+_12756452 | 11.75 |
ENSRNOT00000001382
ENSRNOT00000092462 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr4_-_157304653 | 11.74 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr10_-_56506446 | 11.68 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr2_+_92574038 | 11.64 |
ENSRNOT00000089422
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr10_-_55515885 | 11.64 |
ENSRNOT00000005667
|
Odf4
|
outer dense fiber of sperm tails 4 |
| chr10_+_70242874 | 11.56 |
ENSRNOT00000010924
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr9_+_16529579 | 11.54 |
ENSRNOT00000067894
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chr6_+_72092194 | 11.43 |
ENSRNOT00000005670
|
G2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr9_+_16530074 | 11.43 |
ENSRNOT00000091565
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chr10_-_97750679 | 11.43 |
ENSRNOT00000078059
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr3_-_153321352 | 11.33 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr1_+_162890475 | 11.26 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr2_-_144323254 | 11.23 |
ENSRNOT00000018780
|
Sertm1
|
serine-rich and transmembrane domain containing 1 |
| chr1_-_88723973 | 11.19 |
ENSRNOT00000082555
|
NEWGENE_1306714
|
WD repeat domain 62-like 1 |
| chr4_+_57204944 | 11.11 |
ENSRNOT00000085027
|
Strip2
|
striatin interacting protein 2 |
| chr2_+_92549479 | 11.04 |
ENSRNOT00000082912
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr1_+_81683463 | 11.03 |
ENSRNOT00000057600
ENSRNOT00000034553 |
Ceacam6
|
carcinoembryonic antigen-related cell adhesion molecule 6 |
| chr1_+_100832324 | 11.02 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr1_+_82169620 | 11.02 |
ENSRNOT00000088955
ENSRNOT00000068251 |
Prr19
|
proline rich 19 |
| chr13_+_95887708 | 11.01 |
ENSRNOT00000029081
|
LOC689766
|
hypothetical protein LOC689766 |
| chr1_+_253221812 | 10.97 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr9_+_88964525 | 10.95 |
ENSRNOT00000068236
|
Daw1
|
dynein assembly factor with WD repeats 1 |
| chr2_+_240527130 | 10.94 |
ENSRNOT00000031348
|
Slc9b1
|
solute carrier family 9 member B1 |
| chr15_-_29548400 | 10.91 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr2_-_220101791 | 10.90 |
ENSRNOT00000022905
|
Plppr5
|
phospholipid phosphatase related 5 |
| chr7_-_129947991 | 10.89 |
ENSRNOT00000064326
|
AABR07058658.1
|
|
| chr3_+_122928964 | 10.86 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr8_-_111116014 | 10.79 |
ENSRNOT00000056531
|
Cep63
|
centrosomal protein 63 |
| chr2_+_189454340 | 10.78 |
ENSRNOT00000087063
|
Nup210l
|
nucleoporin 210-like |
| chr12_+_45319501 | 10.73 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr13_-_51955735 | 10.71 |
ENSRNOT00000007597
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr3_-_123171875 | 10.67 |
ENSRNOT00000028834
|
Ubox5
|
U-box domain containing 5 |
| chr6_+_43644830 | 10.66 |
ENSRNOT00000086798
|
Taf1b
|
TATA-box binding protein associated factor, RNA polymerase I subunit B |
| chr3_-_9636058 | 10.60 |
ENSRNOT00000024953
|
RGD1311084
|
similar to 1700113K14Rik protein |
| chr9_+_20241062 | 10.58 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr2_+_197682000 | 10.57 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr14_+_17534412 | 10.55 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr4_+_61420009 | 10.52 |
ENSRNOT00000082175
ENSRNOT00000011692 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr5_-_171327450 | 10.52 |
ENSRNOT00000088710
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr17_+_81798756 | 10.52 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr6_+_22296128 | 10.49 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_+_219390523 | 10.47 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr17_+_5311274 | 10.44 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr2_-_228665279 | 10.42 |
ENSRNOT00000055648
|
RGD1306519
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr2_-_265300868 | 10.39 |
ENSRNOT00000066024
ENSRNOT00000016073 ENSRNOT00000033502 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr3_+_148837814 | 10.36 |
ENSRNOT00000091808
|
Asxl1
|
additional sex combs like 1 |
| chr8_+_23398030 | 10.34 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr10_-_55560422 | 10.31 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr10_-_12916784 | 10.30 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr1_-_81144024 | 10.30 |
ENSRNOT00000088431
|
Zfp94
|
zinc finger protein 94 |
| chr1_+_101603222 | 10.29 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr16_-_49453394 | 10.27 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr3_+_21555341 | 10.23 |
ENSRNOT00000030332
|
AABR07051793.1
|
|
| chr10_+_57239993 | 10.23 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr2_+_231884337 | 10.22 |
ENSRNOT00000014695
|
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr1_-_66212418 | 10.21 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr16_+_66230011 | 10.21 |
ENSRNOT00000074550
|
LOC689479
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr1_+_199019289 | 10.20 |
ENSRNOT00000087142
|
Phkg2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr10_-_82492918 | 10.20 |
ENSRNOT00000066895
|
LOC100364194
|
transmembrane protein 92-like |
| chr5_+_8916633 | 10.15 |
ENSRNOT00000066395
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr12_-_24365324 | 10.06 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr3_+_161290391 | 10.01 |
ENSRNOT00000021070
|
Zswim1
|
zinc finger, SWIM-type containing 1 |
| chr3_+_163552166 | 10.01 |
ENSRNOT00000049082
|
Tp53rk
|
TP53 regulating kinase |
| chr10_+_62981297 | 9.98 |
ENSRNOT00000031618
|
Efcab5
|
EF-hand calcium binding domain 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx2_Otx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 38.0 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 11.4 | 57.0 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 9.0 | 27.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 9.0 | 26.9 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 7.0 | 35.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 5.3 | 21.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 4.8 | 19.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 4.8 | 23.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 4.5 | 18.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 4.4 | 13.2 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 4.2 | 16.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 3.7 | 11.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 3.6 | 18.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 3.6 | 39.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 3.6 | 28.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 3.5 | 10.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 3.5 | 10.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 3.4 | 10.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 3.2 | 9.7 | GO:0032258 | CVT pathway(GO:0032258) |
| 3.2 | 9.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 3.2 | 25.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 3.1 | 12.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 3.0 | 39.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 2.9 | 8.8 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 2.9 | 8.7 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 2.8 | 14.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 2.7 | 8.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 2.7 | 10.7 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 2.6 | 40.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 2.5 | 7.6 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 2.4 | 7.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 2.3 | 9.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 2.3 | 6.9 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 2.3 | 6.9 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 2.2 | 13.2 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 2.2 | 8.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 2.2 | 8.7 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 2.2 | 8.7 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 2.1 | 12.9 | GO:0007343 | egg activation(GO:0007343) |
| 2.1 | 6.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 2.1 | 8.5 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 2.1 | 6.3 | GO:0043366 | beta selection(GO:0043366) |
| 2.1 | 14.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 2.1 | 23.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 2.1 | 33.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 2.0 | 14.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 2.0 | 12.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 2.0 | 21.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.9 | 5.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.9 | 7.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 1.9 | 13.5 | GO:0046959 | habituation(GO:0046959) |
| 1.9 | 11.3 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.8 | 7.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.8 | 21.1 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 1.7 | 5.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 1.7 | 13.7 | GO:0015808 | L-alanine transport(GO:0015808) |
| 1.7 | 8.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.7 | 1.7 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 1.7 | 5.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.6 | 9.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.6 | 3.2 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 1.6 | 4.8 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 1.6 | 4.7 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.6 | 4.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 1.6 | 6.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.5 | 10.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.5 | 22.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 1.5 | 8.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.5 | 8.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.4 | 14.5 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 1.4 | 2.8 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 1.4 | 4.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.4 | 12.6 | GO:0015866 | ADP transport(GO:0015866) |
| 1.4 | 9.8 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 1.4 | 5.6 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.4 | 8.3 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 1.4 | 20.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 1.4 | 4.1 | GO:0015692 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 1.3 | 22.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.3 | 2.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.3 | 9.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 1.3 | 6.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 1.3 | 16.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.2 | 5.9 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 1.2 | 3.5 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 1.2 | 3.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.1 | 10.3 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 1.1 | 10.3 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 1.1 | 3.4 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 1.1 | 4.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 1.1 | 32.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 1.1 | 3.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.1 | 41.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 1.1 | 3.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 1.1 | 5.3 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 5.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.0 | 10.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 1.0 | 7.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 1.0 | 3.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 4.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 1.0 | 15.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.0 | 16.9 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 1.0 | 2.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 1.0 | 3.9 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.9 | 2.8 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.9 | 4.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.9 | 2.8 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.9 | 7.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.9 | 13.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.9 | 7.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.9 | 4.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.9 | 10.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.9 | 4.4 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.9 | 7.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.9 | 6.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.9 | 2.6 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.8 | 2.5 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.8 | 13.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.8 | 7.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.8 | 3.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.8 | 2.4 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.8 | 15.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.8 | 5.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 9.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.8 | 7.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.8 | 3.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.8 | 1.5 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.8 | 2.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.8 | 3.8 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.7 | 15.0 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.7 | 3.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.7 | 1.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.7 | 2.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.7 | 4.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.7 | 2.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.7 | 5.8 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.7 | 7.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.7 | 3.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.7 | 2.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.7 | 2.8 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.7 | 3.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.7 | 6.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.7 | 4.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.7 | 6.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 2.0 | GO:0071049 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.7 | 2.0 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 0.7 | 5.9 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.7 | 2.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.6 | 7.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 0.6 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.6 | 3.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.6 | 1.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.6 | 3.2 | GO:0009838 | abscission(GO:0009838) |
| 0.6 | 13.9 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.6 | 16.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.6 | 3.7 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 12.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.6 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.6 | 3.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.6 | 7.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 4.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.6 | 3.0 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.6 | 3.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.6 | 3.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.6 | 12.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.6 | 2.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.6 | 2.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 4.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.6 | 3.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.6 | 1.8 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.6 | 6.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 2.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.6 | 8.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 5.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.6 | 4.5 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.6 | 12.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.6 | 3.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.6 | 9.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.6 | 6.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.5 | 2.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 3.3 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 4.9 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.5 | 3.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.5 | 4.3 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.5 | 27.8 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.5 | 2.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.5 | 8.9 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.5 | 9.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.5 | 2.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.5 | 7.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 3.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.5 | 1.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.5 | 1.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.5 | 8.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.5 | 3.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 4.5 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.5 | 4.5 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.5 | 6.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.5 | 8.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.5 | 7.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.5 | 1.4 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.5 | 6.7 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.5 | 1.9 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.5 | 7.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.5 | 3.8 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.5 | 2.8 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.5 | 11.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.5 | 4.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.4 | 3.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.4 | 2.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.4 | 5.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.4 | 10.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.4 | 4.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 3.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.4 | 2.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 1.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 2.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) response to amino acid starvation(GO:1990928) |
| 0.4 | 11.0 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.4 | 1.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.4 | 1.2 | GO:0097116 | postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.4 | 4.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 2.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 | 1.2 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.4 | 2.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.4 | 2.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 0.4 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 | 15.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.4 | 6.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.4 | 2.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 4.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 1.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.4 | 3.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 8.5 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.4 | 0.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.4 | 4.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 2.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 1.1 | GO:0046099 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 0.4 | 4.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 1.4 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 1.4 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.3 | 4.5 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.3 | 144.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.3 | 21.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 11.9 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.3 | 2.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 1.0 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 | 1.0 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 0.3 | 19.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.3 | 5.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 4.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.3 | 1.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 4.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.3 | 7.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 1.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 2.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 2.8 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.3 | 3.7 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.3 | 2.7 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 5.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 2.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.3 | 8.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 0.8 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.3 | 5.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.3 | 13.7 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.3 | 45.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.3 | 5.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 2.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 1.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 1.6 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 11.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 2.1 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 1.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 6.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 5.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 2.9 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 2.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 2.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 2.6 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.2 | 4.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 4.6 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 4.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 1.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 2.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 12.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 4.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 1.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 5.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 7.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 4.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 3.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 6.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 1.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 3.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 5.5 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.2 | 5.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 1.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 3.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 3.5 | GO:0009629 | response to gravity(GO:0009629) |
| 0.2 | 3.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.2 | 3.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 2.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 0.7 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 2.4 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 | 1.8 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.2 | 3.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.2 | 5.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 6.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 1.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 2.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 1.0 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 0.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 3.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 1.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 5.0 | GO:1902117 | positive regulation of organelle assembly(GO:1902117) |
| 0.2 | 1.6 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 3.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 11.6 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.2 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.2 | 0.9 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 2.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.3 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 4.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 3.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 2.1 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 14.0 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.6 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 9.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 3.2 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 2.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.3 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 18.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 0.7 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 2.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 4.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 5.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 0.4 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 1.0 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 6.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 9.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 2.5 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 3.6 | GO:0009411 | response to UV(GO:0009411) |
| 0.1 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.4 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 2.0 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 1.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 3.6 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 2.6 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.3 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 7.4 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.1 | 1.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 1.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 1.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 4.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.4 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.2 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 3.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 38.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 5.3 | 15.8 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 4.9 | 29.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 4.5 | 4.5 | GO:0044317 | rod spherule(GO:0044317) |
| 4.3 | 30.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 3.6 | 18.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 3.3 | 30.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 3.2 | 9.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 2.9 | 41.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 2.8 | 16.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.6 | 17.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 2.5 | 10.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 2.5 | 5.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 2.5 | 12.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 2.3 | 7.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 2.3 | 9.2 | GO:0033503 | HULC complex(GO:0033503) |
| 2.1 | 6.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.1 | 10.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 2.1 | 6.4 | GO:0032301 | recombination nodule(GO:0005713) MutSalpha complex(GO:0032301) |
| 2.1 | 12.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 2.1 | 20.7 | GO:0044615 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 2.0 | 8.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.9 | 7.7 | GO:0072487 | MSL complex(GO:0072487) |
| 1.9 | 20.7 | GO:0002177 | manchette(GO:0002177) |
| 1.9 | 33.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.8 | 5.5 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.6 | 4.8 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 1.5 | 13.7 | GO:0071546 | pi-body(GO:0071546) |
| 1.5 | 8.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.5 | 4.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.5 | 4.4 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.4 | 4.3 | GO:0000802 | transverse filament(GO:0000802) |
| 1.4 | 4.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 1.4 | 13.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.3 | 6.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.3 | 10.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 1.3 | 5.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.2 | 12.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.2 | 14.7 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 1.1 | 6.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.1 | 50.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 1.1 | 11.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.1 | 10.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.0 | 7.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 1.0 | 4.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.0 | 9.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 1.0 | 2.9 | GO:0000811 | GINS complex(GO:0000811) |
| 1.0 | 14.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.0 | 5.8 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.9 | 14.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.9 | 4.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.9 | 2.6 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.9 | 10.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.9 | 8.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.9 | 11.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.9 | 3.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 94.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.8 | 11.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.8 | 3.3 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.8 | 4.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.8 | 10.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.8 | 12.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.8 | 2.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.8 | 12.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 2.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.7 | 6.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 7.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.7 | 4.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.7 | 6.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 21.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 4.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.6 | 5.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 3.8 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.6 | 14.6 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.6 | 76.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.6 | 3.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.6 | 66.6 | GO:0005814 | centriole(GO:0005814) |
| 0.6 | 5.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.6 | 5.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 6.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 3.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.6 | 2.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 2.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 10.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.5 | 5.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 21.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.5 | 4.0 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.5 | 5.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.5 | 3.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 2.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.5 | 19.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 4.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.4 | 4.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 4.8 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.4 | 2.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 54.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.4 | 5.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.4 | 2.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 10.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 18.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.4 | 3.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 24.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.4 | 1.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.4 | 15.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 5.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.4 | 2.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 5.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.4 | 8.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 16.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.4 | 1.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 6.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 3.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 5.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 1.4 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.3 | 5.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 1.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.3 | 2.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 2.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 2.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 2.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 2.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 5.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 3.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.3 | 6.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 4.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 18.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 11.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 4.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 1.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 3.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 2.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 0.7 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 1.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.7 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 1.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 2.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 3.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 8.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 5.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 6.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 2.1 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.2 | 1.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 4.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 2.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 12.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 23.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 5.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 14.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 4.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 3.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 10.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.2 | 3.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 6.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 4.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 2.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 9.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 7.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 33.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 21.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 14.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 8.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 5.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 21.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 43.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 2.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.4 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 64.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 5.9 | 35.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 4.9 | 14.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 4.7 | 14.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 4.4 | 13.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 4.2 | 29.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 3.7 | 11.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 3.5 | 20.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 3.2 | 9.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 3.1 | 50.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.8 | 8.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 2.7 | 13.7 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 2.7 | 8.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 2.7 | 42.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 2.6 | 7.8 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 2.6 | 10.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.5 | 7.6 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 2.4 | 9.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.3 | 16.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 2.3 | 35.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 2.2 | 21.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.1 | 23.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 2.1 | 16.9 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 2.0 | 7.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.9 | 5.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.8 | 53.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 1.8 | 5.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.8 | 10.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.8 | 5.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.7 | 8.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.7 | 19.0 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.6 | 13.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.6 | 6.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.6 | 12.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.5 | 7.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.5 | 4.4 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.5 | 4.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 1.5 | 8.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.4 | 8.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.4 | 11.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.4 | 11.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.4 | 12.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.4 | 4.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 1.4 | 5.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.3 | 26.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.3 | 6.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.3 | 10.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.2 | 13.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.2 | 26.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 1.2 | 5.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.2 | 4.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.2 | 13.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 1.1 | 10.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.1 | 16.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.1 | 7.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 4.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.0 | 3.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.0 | 5.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 1.0 | 4.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.0 | 3.9 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 1.0 | 15.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.0 | 3.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.9 | 2.8 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.9 | 7.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.9 | 4.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.9 | 3.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.9 | 2.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.9 | 2.8 | GO:0001221 | transcription cofactor binding(GO:0001221) transcription corepressor binding(GO:0001222) |
| 0.9 | 4.6 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.9 | 3.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.9 | 2.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.9 | 10.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.9 | 5.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.9 | 9.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.9 | 2.6 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.8 | 5.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 7.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.8 | 24.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.8 | 3.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.8 | 16.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.8 | 15.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.8 | 2.3 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.8 | 9.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 8.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.7 | 9.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.7 | 16.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.7 | 5.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.7 | 9.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 7.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 3.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.7 | 2.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.7 | 2.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.7 | 20.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.7 | 8.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 5.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.7 | 2.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.6 | 7.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.6 | 6.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.6 | 1.9 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.6 | 20.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.6 | 3.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 12.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.6 | 4.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.6 | 3.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.6 | 4.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.6 | 7.4 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.6 | 2.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.6 | 13.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.6 | 14.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.5 | 2.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.5 | 2.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 3.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 13.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 1.6 | GO:0035596 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.5 | 3.1 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.5 | 1.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 9.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.5 | 2.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.5 | 12.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.5 | 8.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 2.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.5 | 2.4 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 5.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 15.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.5 | 11.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.5 | 7.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 1.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.4 | 1.3 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.4 | 6.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 8.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 13.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 6.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 2.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 29.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 2.9 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.4 | 3.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 3.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.4 | 4.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 2.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.4 | 1.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.4 | 3.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 3.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 10.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 1.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 22.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.4 | 1.5 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.4 | 6.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 6.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.4 | 4.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.4 | 2.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.4 | 2.9 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.4 | 12.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.4 | 24.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 2.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 3.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.4 | 1.4 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.3 | 2.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 6.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 4.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 1.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.3 | 3.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 19.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.3 | 3.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 7.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 6.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 2.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 3.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 5.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 2.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 2.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 3.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 1.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 1.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 6.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 8.8 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.3 | 8.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 3.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.7 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 14.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 2.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 1.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 3.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 4.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 6.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 5.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 13.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 25.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 6.3 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.2 | 3.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 12.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 50.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 5.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 2.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 6.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 0.4 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.2 | 3.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 2.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.6 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.2 | 10.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 8.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 3.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 5.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 2.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 3.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.2 | 2.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 17.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 4.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 3.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 21.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 9.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 3.2 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.2 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 35.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 6.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 1.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 3.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.2 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.2 | 1.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 6.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 3.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.9 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 3.1 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 4.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.9 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.1 | 5.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 1.2 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.1 | 4.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 11.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 16.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 3.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 26.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 1.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 3.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 4.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.6 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 14.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.0 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 1.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 4.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 5.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 64.9 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.8 | 31.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.8 | 30.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.7 | 8.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 20.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.7 | 22.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.7 | 30.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 5.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.7 | 20.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.6 | 23.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 13.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 43.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.5 | 4.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.5 | 9.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.5 | 10.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 26.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 4.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.4 | 7.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 5.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 17.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 14.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 2.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 8.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 7.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 11.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 10.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 1.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 9.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 9.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 10.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 5.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 5.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 32.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 1.9 | 29.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.3 | 20.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.2 | 65.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 1.2 | 11.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.1 | 13.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.0 | 16.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.0 | 7.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 29.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 20.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.9 | 31.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.8 | 8.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.8 | 23.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.8 | 6.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 20.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.7 | 3.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.7 | 21.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.7 | 11.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 13.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.6 | 28.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.6 | 6.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 4.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.5 | 12.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.5 | 8.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.5 | 13.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 4.1 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.5 | 11.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 8.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.5 | 8.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.5 | 14.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 6.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.4 | 13.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 27.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 32.0 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.4 | 4.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.4 | 8.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 13.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.4 | 8.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 9.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 4.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 5.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 11.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 2.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 2.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.3 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 4.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 5.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 39.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 3.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 7.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.2 | 3.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 8.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 4.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 3.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 10.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 22.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 9.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.3 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |