Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
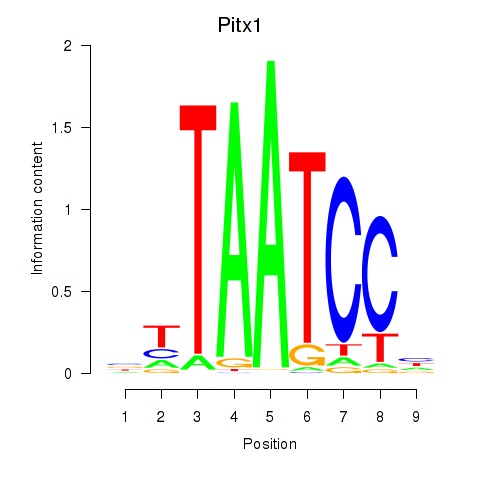
Results for Pitx1
Z-value: 0.90
Transcription factors associated with Pitx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx1
|
ENSRNOG00000011423 | paired-like homeodomain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx1 | rn6_v1_chr17_+_8878270_8878270 | 0.49 | 7.0e-21 | Click! |
Activity profile of Pitx1 motif
Sorted Z-values of Pitx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_224824799 | 44.24 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr10_+_53818818 | 29.09 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr20_+_20378861 | 25.44 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chrX_+_159158194 | 24.12 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr3_+_55910177 | 23.41 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr3_+_113318563 | 22.44 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr3_+_80075991 | 21.39 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr4_-_51199570 | 20.89 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr8_+_33514042 | 18.03 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr3_+_56355431 | 17.88 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr5_-_135025084 | 17.44 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr1_+_137799185 | 17.35 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr4_+_65801505 | 16.03 |
ENSRNOT00000018331
|
Tmem213
|
transmembrane protein 213 |
| chr10_-_88036040 | 16.02 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr3_+_117421604 | 15.93 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr7_-_49741540 | 15.82 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr10_+_104368247 | 14.53 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr18_-_24929091 | 14.40 |
ENSRNOT00000019596
|
Proc
|
protein C |
| chr14_+_84306466 | 14.28 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr5_+_29538380 | 14.25 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr8_-_105462141 | 13.83 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr20_-_5123073 | 13.80 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr8_-_114449956 | 13.49 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr7_+_94795214 | 12.77 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr18_+_56193978 | 11.49 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_+_89008117 | 11.34 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr13_+_85818427 | 11.26 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr3_+_112242270 | 11.25 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr15_+_83707735 | 11.10 |
ENSRNOT00000057843
|
Klf5
|
Kruppel-like factor 5 |
| chr10_-_82887301 | 10.89 |
ENSRNOT00000035894
|
Itga3
|
integrin subunit alpha 3 |
| chr9_+_81816872 | 10.81 |
ENSRNOT00000041407
|
Plcd4
|
phospholipase C, delta 4 |
| chr11_-_35697072 | 10.43 |
ENSRNOT00000039999
|
Erg
|
ERG, ETS transcription factor |
| chr2_-_259382765 | 9.77 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr2_+_45104305 | 9.75 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr8_+_62925357 | 9.61 |
ENSRNOT00000011074
ENSRNOT00000090588 |
Stra6
|
stimulated by retinoic acid 6 |
| chr16_+_2670618 | 9.61 |
ENSRNOT00000030102
|
Il17rd
|
interleukin 17 receptor D |
| chr5_-_6186329 | 9.38 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr4_+_130332076 | 9.30 |
ENSRNOT00000083127
|
Mitf
|
melanogenesis associated transcription factor |
| chr7_-_44771458 | 8.91 |
ENSRNOT00000006007
|
Alx1
|
ALX homeobox 1 |
| chr1_-_166943592 | 8.89 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr6_+_56625650 | 8.80 |
ENSRNOT00000008803
|
Meox2
|
mesenchyme homeobox 2 |
| chr4_-_171591882 | 8.77 |
ENSRNOT00000009328
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chrX_+_51286737 | 8.47 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr10_-_88050622 | 8.31 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chrX_-_25628272 | 8.08 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr7_+_60015998 | 7.67 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr18_-_74542077 | 7.50 |
ENSRNOT00000078519
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr17_-_84830185 | 7.27 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_+_15043955 | 7.17 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr1_-_2846200 | 7.11 |
ENSRNOT00000017688
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr10_-_87407634 | 7.05 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr7_-_143497108 | 6.73 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr2_-_206222248 | 6.70 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr5_+_138300107 | 6.66 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr2_-_147392062 | 6.58 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr1_+_268189277 | 6.43 |
ENSRNOT00000065001
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr9_+_81631409 | 6.29 |
ENSRNOT00000089580
|
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr7_-_117732339 | 6.13 |
ENSRNOT00000092917
ENSRNOT00000020696 |
Foxh1
|
forkhead box H1 |
| chr1_-_256734719 | 5.87 |
ENSRNOT00000021546
ENSRNOT00000089456 |
Myof
|
myoferlin |
| chr5_+_104362971 | 5.76 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr2_+_241909832 | 5.64 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_30276372 | 5.25 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr9_+_46962288 | 5.16 |
ENSRNOT00000082146
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr9_-_30844199 | 4.98 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr3_-_113423473 | 4.93 |
ENSRNOT00000064982
|
Serinc4
|
serine incorporator 4 |
| chr6_+_23337571 | 4.81 |
ENSRNOT00000011832
|
RGD1304963
|
similar to hypothetical protein MGC38716 |
| chr10_-_55851235 | 4.62 |
ENSRNOT00000010790
|
Gucy2d
|
guanylate cyclase 2D, retinal |
| chr4_-_16669368 | 4.59 |
ENSRNOT00000007608
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr1_+_107344904 | 4.26 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr2_+_225645568 | 4.15 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr2_-_60657712 | 3.70 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr20_-_7943575 | 3.67 |
ENSRNOT00000066897
|
Tulp1
|
tubby like protein 1 |
| chr5_+_21830882 | 3.56 |
ENSRNOT00000008901
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr7_-_11648322 | 3.52 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr8_-_55171718 | 3.33 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr2_-_45480798 | 3.30 |
ENSRNOT00000066764
|
Snx18
|
sorting nexin 18 |
| chr11_+_30550141 | 3.20 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr12_+_12374790 | 3.17 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr11_-_69201380 | 3.13 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chrM_+_8599 | 3.11 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr1_-_278042312 | 3.10 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_+_145174876 | 3.10 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr1_+_99648658 | 3.07 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr3_-_91195981 | 3.04 |
ENSRNOT00000056935
|
RGD1309730
|
similar to RIKEN cDNA B230118H07 |
| chr3_-_50120392 | 3.01 |
ENSRNOT00000006175
|
Fign
|
fidgetin, microtubule severing factor |
| chr7_-_70476340 | 2.98 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chr4_-_125929002 | 2.94 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_+_3662763 | 2.90 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr10_-_63176463 | 2.84 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr15_+_3938075 | 2.68 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_+_220854403 | 2.67 |
ENSRNOT00000080520
ENSRNOT00000052315 ENSRNOT00000078619 |
Efemp2
|
EGF-containing fibulin-like extracellular matrix protein 2 |
| chr3_+_113423693 | 2.55 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr18_+_71395830 | 2.55 |
ENSRNOT00000024831
|
Smad7
|
SMAD family member 7 |
| chr4_+_155532109 | 2.50 |
ENSRNOT00000077245
|
Nanog
|
Nanog homeobox |
| chr1_+_124983452 | 2.39 |
ENSRNOT00000021262
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_-_210282352 | 2.39 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr19_-_38120578 | 2.32 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr16_+_49185560 | 2.25 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr3_-_44086006 | 2.16 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr1_-_24191908 | 2.15 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_-_108372068 | 2.14 |
ENSRNOT00000088116
ENSRNOT00000091143 ENSRNOT00000085886 |
RGD1305110
|
similar to KIAA1841 protein |
| chr8_+_42511520 | 2.14 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chr1_-_66254989 | 2.07 |
ENSRNOT00000025981
ENSRNOT00000079249 |
Zfp606
|
zinc finger protein 606 |
| chr9_-_121972055 | 2.01 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr6_-_103949275 | 2.00 |
ENSRNOT00000047825
|
AABR07065012.1
|
|
| chr4_+_30313102 | 1.98 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr11_+_57207656 | 1.79 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_-_12916784 | 1.79 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr1_-_4011161 | 1.71 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr3_-_76718684 | 1.69 |
ENSRNOT00000050173
|
Olr631
|
olfactory receptor 631 |
| chr3_+_97723901 | 1.66 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr8_-_42381997 | 1.55 |
ENSRNOT00000052324
|
LOC100912580
|
olfactory receptor 8B3-like |
| chr4_+_1535100 | 1.49 |
ENSRNOT00000074575
|
LOC100912488
|
olfactory receptor 147-like |
| chr3_-_113423149 | 1.49 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr2_+_203768729 | 1.44 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_-_227067915 | 1.39 |
ENSRNOT00000067235
|
Ms4a18
|
membrane spanning 4-domains A18 |
| chr12_+_47179664 | 1.32 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr3_-_74631883 | 1.26 |
ENSRNOT00000013215
|
Olr539
|
olfactory receptor 539 |
| chr12_+_13097269 | 1.20 |
ENSRNOT00000087094
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr10_-_55560422 | 1.16 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr18_-_37096132 | 0.97 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr20_-_8574082 | 0.94 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr11_-_80981415 | 0.93 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr4_+_72480540 | 0.92 |
ENSRNOT00000007163
|
Olr812
|
olfactory receptor 812 |
| chr16_+_23317953 | 0.82 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr18_+_56544652 | 0.76 |
ENSRNOT00000024171
|
Pde6a
|
phosphodiesterase 6A |
| chr8_-_68525911 | 0.73 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr9_-_23454316 | 0.66 |
ENSRNOT00000072826
|
Cyp2ac1
|
cytochrome P450, family 2, subfamily ac, polypeptide 1 |
| chr11_+_81796891 | 0.66 |
ENSRNOT00000058402
|
Crygs
|
crystallin, gamma S |
| chr7_+_120176530 | 0.64 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr3_-_73049251 | 0.47 |
ENSRNOT00000048041
|
Olr450
|
olfactory receptor 450 |
| chr10_-_85435016 | 0.45 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr6_-_103807838 | 0.43 |
ENSRNOT00000058328
|
AABR07064998.1
|
|
| chr12_-_35979193 | 0.39 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr8_+_41936959 | 0.38 |
ENSRNOT00000042601
|
Olr1222
|
olfactory receptor 1222 |
| chr12_-_39697465 | 0.35 |
ENSRNOT00000078683
|
Vps29
|
VPS29 retromer complex component |
| chr6_-_104409005 | 0.31 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr1_-_8885967 | 0.29 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr5_+_135997052 | 0.27 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr1_-_216744066 | 0.25 |
ENSRNOT00000087961
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr7_+_5593735 | 0.22 |
ENSRNOT00000042179
|
Olr917
|
olfactory receptor 917 |
| chr19_-_43215077 | 0.22 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr3_+_73161632 | 0.20 |
ENSRNOT00000077865
|
Olr458
|
olfactory receptor 458 |
| chr2_+_46066780 | 0.20 |
ENSRNOT00000073226
|
LOC108350225
|
olfactory receptor 8B8-like |
| chr20_+_28989491 | 0.16 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr16_+_14319777 | 0.15 |
ENSRNOT00000038771
|
Lrit2
|
leucine-rich repeat, Ig-like and transmembrane domains 2 |
| chr2_+_128461224 | 0.15 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr5_+_133864798 | 0.15 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr5_+_49311030 | 0.08 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr7_-_120490061 | 0.06 |
ENSRNOT00000016247
|
Slc16a8
|
solute carrier family 16 member 8 |
| chr2_+_60966789 | 0.02 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr16_+_10277775 | 0.02 |
ENSRNOT00000090679
|
Rbp3
|
retinol binding protein 3 |
| chr1_+_256955652 | 0.01 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 44.2 | GO:0031427 | response to methotrexate(GO:0031427) |
| 6.4 | 25.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 6.3 | 19.0 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 6.0 | 18.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 5.3 | 15.9 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.7 | 11.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 3.6 | 14.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 3.6 | 14.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 3.2 | 9.6 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 3.1 | 9.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 3.0 | 8.9 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 2.9 | 8.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 2.9 | 17.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 2.8 | 8.5 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 2.6 | 29.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 2.4 | 9.7 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 2.4 | 14.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 2.3 | 11.5 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 2.0 | 6.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 2.0 | 15.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.9 | 5.6 | GO:1905203 | regulation of connective tissue replacement(GO:1905203) |
| 1.8 | 7.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.8 | 8.8 | GO:0001757 | somite specification(GO:0001757) |
| 1.7 | 10.4 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 1.7 | 5.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.7 | 23.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) regulation of myoblast proliferation(GO:2000291) |
| 1.6 | 9.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 1.4 | 4.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 1.3 | 16.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.3 | 6.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.2 | 7.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.2 | 3.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.1 | 21.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.1 | 8.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.9 | 11.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.9 | 9.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.8 | 14.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.8 | 10.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.8 | 2.4 | GO:0015820 | leucine transport(GO:0015820) |
| 0.8 | 3.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 4.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 8.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.7 | 3.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.6 | 8.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.6 | 9.8 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.6 | 2.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.5 | 24.1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.5 | 6.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 5.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 6.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.4 | 2.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.4 | 11.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.4 | 14.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 2.5 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.4 | 1.8 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.4 | 11.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.3 | 0.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.3 | 6.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.3 | 1.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.3 | 1.3 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.3 | 10.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 8.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 1.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 2.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 2.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 3.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 13.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 2.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 0.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 2.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 16.8 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 1.4 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 3.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 16.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 2.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 1.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 7.8 | GO:0043271 | negative regulation of ion transport(GO:0043271) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 3.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 1.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 4.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 3.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 2.2 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 2.9 | 11.5 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 2.8 | 13.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.5 | 25.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.9 | 29.1 | GO:0032982 | myosin filament(GO:0032982) |
| 1.8 | 23.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.5 | 4.6 | GO:0044317 | rod spherule(GO:0044317) |
| 1.3 | 17.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.0 | 6.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.9 | 5.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 2.4 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.7 | 5.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 7.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 8.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.5 | 8.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 46.3 | GO:0005901 | caveola(GO:0005901) |
| 0.4 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 5.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 24.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 2.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 7.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 3.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 9.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 7.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 11.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 19.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 3.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 9.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 22.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 9.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 26.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 17.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 24.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 14.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 48.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 3.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 9.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 16.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 3.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 44.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 5.3 | 15.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 4.8 | 14.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 4.5 | 18.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 3.6 | 14.2 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 3.2 | 9.7 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 3.2 | 22.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.3 | 9.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.2 | 8.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.1 | 47.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 2.0 | 9.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.9 | 21.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 1.9 | 11.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.7 | 5.2 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.7 | 5.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.4 | 5.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.3 | 6.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.2 | 9.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.1 | 7.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 1.0 | 11.2 | GO:0031432 | titin binding(GO:0031432) |
| 1.0 | 11.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.9 | 2.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.9 | 14.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.8 | 4.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.8 | 3.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 17.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 10.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 8.5 | GO:0050998 | vinculin binding(GO:0017166) nitric-oxide synthase binding(GO:0050998) |
| 0.4 | 2.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 14.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 16.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 7.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 9.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.3 | 10.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 1.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 2.5 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.3 | 22.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 4.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 7.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 0.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 8.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.2 | 3.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 13.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 14.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 7.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 8.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 25.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 10.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 6.5 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 14.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 10.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 14.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 11.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 13.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 14.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 2.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 3.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 9.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 19.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 9.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 5.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 15.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.7 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 44.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.5 | 20.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.4 | 14.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.0 | 37.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 25.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 10.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 11.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.5 | 10.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 12.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 6.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 8.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 7.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 6.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 11.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 6.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 7.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 3.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |