Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
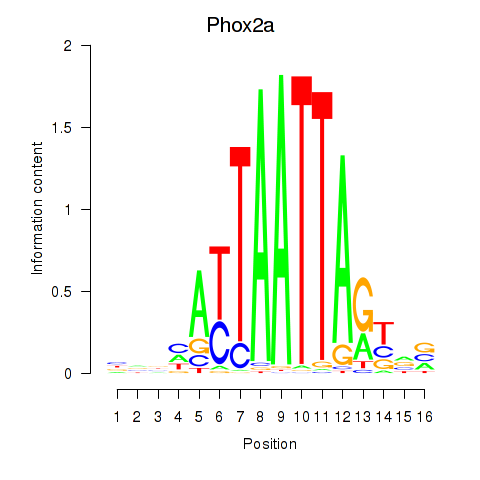
Results for Phox2a
Z-value: 0.33
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSRNOG00000019706 | paired-like homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2a | rn6_v1_chr1_+_166893734_166893734 | -0.43 | 4.7e-16 | Click! |
Activity profile of Phox2a motif
Sorted Z-values of Phox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_84506328 | 10.24 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr14_-_19159923 | 8.24 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr4_+_180291389 | 6.92 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chrX_+_78042859 | 6.38 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr11_+_58624198 | 5.08 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr14_-_84334066 | 4.49 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr1_-_215033460 | 4.37 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr3_-_165537940 | 4.33 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr2_+_113984646 | 4.30 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr17_-_84247038 | 3.80 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr20_-_13994794 | 3.32 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr4_-_18396035 | 2.87 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr13_+_90943255 | 2.76 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr6_+_80108655 | 2.72 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chrX_-_111191932 | 2.68 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr20_+_42966140 | 2.39 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr5_+_36566783 | 2.10 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr3_+_14889510 | 2.08 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr2_-_158156444 | 2.02 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_158097843 | 2.01 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr2_+_239415046 | 1.79 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr2_+_145174876 | 1.70 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr7_-_126382449 | 1.58 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chrM_+_2740 | 1.51 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr1_-_101095594 | 1.38 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr4_-_58250798 | 1.31 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr2_-_170301348 | 1.06 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr5_+_58995249 | 0.89 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr7_-_140291620 | 0.88 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr9_-_30844199 | 0.87 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr2_+_118910587 | 0.87 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr13_-_90405591 | 0.79 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr10_-_88000423 | 0.79 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr7_-_143967484 | 0.66 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr9_+_15166118 | 0.54 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr7_+_9294709 | 0.43 |
ENSRNOT00000048396
|
Olr1065
|
olfactory receptor 1065 |
| chr1_-_276228574 | 0.32 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr16_-_29936307 | 0.27 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr7_-_69982592 | 0.25 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr12_-_2174131 | 0.22 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr2_+_252090669 | 0.19 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_-_190370499 | 0.19 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr12_-_35979193 | 0.05 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Phox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.7 | 2.9 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.5 | 2.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.5 | 8.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.5 | 5.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 2.0 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.4 | 3.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 10.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.3 | 2.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 4.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 0.8 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 4.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 6.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 4.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.9 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.1 | 1.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 4.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 2.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.6 | 5.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.6 | 2.4 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.4 | 2.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 6.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 10.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 4.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 5.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.6 | 3.3 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 2.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 10.2 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 4.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 8.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 8.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |