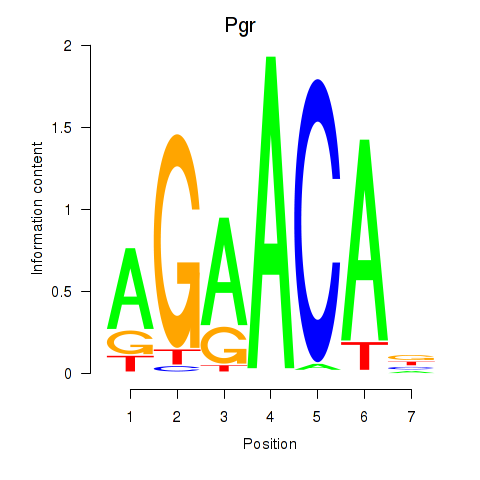
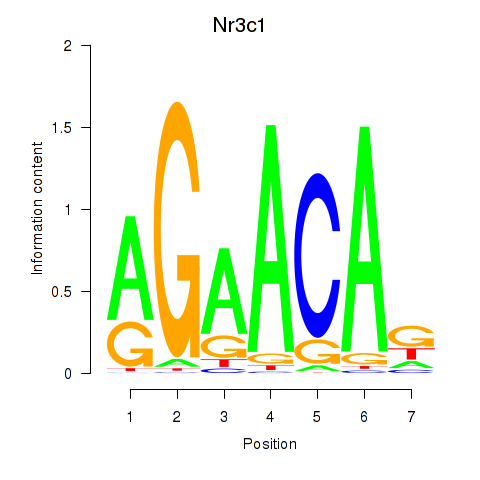
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pgr_Nr3c1
Z-value: 0.32
Transcription factors associated with Pgr_Nr3c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pgr
|
ENSRNOG00000006831 | progesterone receptor |
|
Nr3c1
|
ENSRNOG00000014096 | nuclear receptor subfamily 3, group C, member 1 |
|
Nr3c1
|
ENSRNOG00000048800 | nuclear receptor subfamily 3, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c1 | rn6_v1_chr18_-_31749647_31749647 | 0.45 | 9.8e-18 | Click! |
| Pgr | rn6_v1_chr8_+_7128656_7128656 | -0.08 | 1.7e-01 | Click! |
Activity profile of Pgr_Nr3c1 motif
Sorted Z-values of Pgr_Nr3c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_52624878 | 12.51 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr10_+_92628356 | 11.17 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr10_+_5930298 | 8.11 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr7_+_38819771 | 5.29 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr3_+_129599353 | 4.41 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
| chr2_-_184993341 | 4.30 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr10_-_83898527 | 4.10 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_-_42127678 | 3.41 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_+_184230459 | 3.24 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr3_+_177310753 | 3.24 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr10_+_5199226 | 3.23 |
ENSRNOT00000003544
|
Dexi
|
dexamethasone-induced transcript |
| chr5_-_65073012 | 3.23 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr9_+_17705212 | 2.94 |
ENSRNOT00000026728
|
Tmem63b
|
transmembrane protein 63B |
| chr4_+_71621729 | 2.89 |
ENSRNOT00000022275
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr2_+_116970344 | 2.83 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr6_+_18970564 | 2.69 |
ENSRNOT00000090121
ENSRNOT00000030803 |
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr10_-_60542410 | 2.44 |
ENSRNOT00000082426
|
Olr1498
|
olfactory receptor 1498 |
| chr19_+_46733633 | 2.12 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chr15_+_58016238 | 1.87 |
ENSRNOT00000075786
|
Kctd4
|
potassium channel tetramerization domain containing 4 |
| chr16_+_3293599 | 1.62 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr2_+_205525204 | 1.55 |
ENSRNOT00000091210
|
Csde1
|
cold shock domain containing E1 |
| chr1_-_171028743 | 1.14 |
ENSRNOT00000026488
|
Olr223
|
olfactory receptor 223 |
| chr10_-_60309306 | 0.92 |
ENSRNOT00000083439
|
Olr1486
|
olfactory receptor 1486 |
| chr3_-_76114902 | 0.86 |
ENSRNOT00000071564
|
LOC102555599
|
olfactory receptor 5L1-like |
| chr3_+_110508403 | 0.84 |
ENSRNOT00000079959
|
AABR07053500.2
|
|
| chr7_-_104556419 | 0.75 |
ENSRNOT00000090784
|
Fam49b
|
family with sequence similarity 49, member B |
| chr8_+_42817108 | 0.74 |
ENSRNOT00000072645
|
LOC100910120
|
olfactory receptor 8D1-like |
| chr4_+_72239942 | 0.72 |
ENSRNOT00000048423
|
Olr807
|
olfactory receptor 807 |
| chr11_+_43143882 | 0.59 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr7_+_42269784 | 0.47 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr11_-_45616429 | 0.42 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
| chr11_-_45667254 | 0.20 |
ENSRNOT00000072466
|
Olr1535
|
olfactory receptor 1535 |
| chr10_+_60043848 | 0.16 |
ENSRNOT00000080593
|
AABR07029918.1
|
|
| chr11_+_43340505 | 0.12 |
ENSRNOT00000043299
|
Olr1541
|
olfactory receptor 1541 |
| chr8_+_1459526 | 0.07 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pgr_Nr3c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.5 | 4.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.1 | 3.4 | GO:0071282 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) cellular response to iron(II) ion(GO:0071282) |
| 1.0 | 12.5 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.9 | 5.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 11.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 3.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 1.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 0.7 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 3.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 4.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.5 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 12.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.9 | 4.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.7 | 8.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 5.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 3.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 11.2 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 1.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 3.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 11.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.7 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 4.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 4.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 5.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 12.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 5.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 23.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 8.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.4 | 5.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 4.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 4.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |