Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pbx2
Z-value: 0.85
Transcription factors associated with Pbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pbx2
|
ENSRNOG00000000440 | PBX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx2 | rn6_v1_chr20_+_4357733_4357733 | 0.24 | 1.8e-05 | Click! |
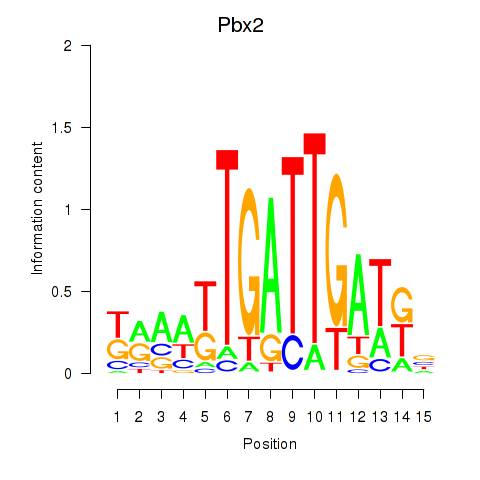
Activity profile of Pbx2 motif
Sorted Z-values of Pbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_201620642 | 52.65 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr7_+_125288081 | 43.02 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr4_+_98337367 | 35.58 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr11_+_85430400 | 32.84 |
ENSRNOT00000083198
|
AABR07034729.1
|
|
| chr9_-_14706557 | 28.61 |
ENSRNOT00000048975
|
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr11_+_85263536 | 25.55 |
ENSRNOT00000046465
|
AABR07034729.1
|
|
| chr1_-_197821936 | 24.93 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr4_+_101639641 | 23.08 |
ENSRNOT00000058282
|
AABR07060952.1
|
|
| chr4_+_102489916 | 22.80 |
ENSRNOT00000082031
|
AABR07061001.1
|
|
| chr9_-_23493081 | 21.84 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr4_+_156752082 | 20.77 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr13_-_55173692 | 20.04 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_88217188 | 20.03 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chrX_+_80359555 | 19.53 |
ENSRNOT00000030692
|
LOC681325
|
hypothetical protein LOC681325 |
| chr4_-_163849618 | 19.28 |
ENSRNOT00000086363
ENSRNOT00000077637 |
Ly49si1
|
immunoreceptor Ly49si1 |
| chr15_+_57241968 | 18.95 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_-_209537087 | 18.82 |
ENSRNOT00000024344
|
Cd53
|
Cd53 molecule |
| chr4_-_163762434 | 16.59 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr10_-_34242985 | 16.47 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr4_+_69138525 | 16.19 |
ENSRNOT00000073589
|
Trbv1
|
T cell receptor beta, variable 1 |
| chr13_+_101381510 | 16.09 |
ENSRNOT00000004821
|
Tlr5
|
toll-like receptor 5 |
| chr3_-_113405829 | 15.07 |
ENSRNOT00000036823
|
Ell3
|
elongation factor for RNA polymerase II 3 |
| chr2_+_183674522 | 14.44 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr4_+_102403451 | 14.24 |
ENSRNOT00000071605
|
AABR07060995.1
|
|
| chr4_-_102124609 | 13.90 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr5_+_133865331 | 13.82 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr11_+_85992696 | 13.50 |
ENSRNOT00000084433
|
AABR07034736.1
|
|
| chr2_-_218658761 | 13.39 |
ENSRNOT00000018318
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr4_+_163174487 | 13.25 |
ENSRNOT00000088108
|
Clec9a
|
C-type lectin domain family 9, member A |
| chr9_+_67699379 | 13.10 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr1_-_98521551 | 13.06 |
ENSRNOT00000081922
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr19_-_19727081 | 13.01 |
ENSRNOT00000020700
|
Adcy7
|
adenylate cyclase 7 |
| chr1_+_219390523 | 12.88 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr7_-_81592206 | 12.87 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr2_+_188449210 | 11.93 |
ENSRNOT00000027700
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr4_-_34282351 | 11.74 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chr8_-_124399494 | 11.72 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr1_-_133559975 | 11.51 |
ENSRNOT00000046645
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr4_-_163214678 | 10.53 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr2_-_203350736 | 10.36 |
ENSRNOT00000090115
ENSRNOT00000084516 |
Ttf2
|
transcription termination factor 2 |
| chr8_+_130538651 | 10.27 |
ENSRNOT00000026343
|
Ackr2
|
atypical chemokine receptor 2 |
| chr6_+_137323713 | 10.01 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr4_-_164691405 | 9.60 |
ENSRNOT00000090979
ENSRNOT00000091932 ENSRNOT00000078219 |
Ly49s4
Ly49i2
|
Ly49 stimulatory receptor 4 Ly49 inhibitory receptor 2 |
| chr3_-_46457201 | 9.32 |
ENSRNOT00000010683
|
Ly75
|
lymphocyte antigen 75 |
| chr7_-_14189688 | 9.32 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr18_+_55666027 | 9.17 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr17_-_43614844 | 9.07 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr13_+_82496022 | 9.06 |
ENSRNOT00000080759
|
F5
|
coagulation factor V |
| chr9_-_20154077 | 8.57 |
ENSRNOT00000082904
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr1_+_84280945 | 8.52 |
ENSRNOT00000057190
|
Sertad3
|
SERTA domain containing 3 |
| chr4_+_34282625 | 8.27 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr7_+_144064931 | 7.65 |
ENSRNOT00000048504
|
Prr13
|
proline rich 13 |
| chr1_-_218810118 | 7.54 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr8_+_2635851 | 7.34 |
ENSRNOT00000061973
|
Casp4
|
caspase 4 |
| chrX_+_124894466 | 7.28 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr15_-_24199341 | 7.02 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr5_+_133864798 | 6.95 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr17_+_43673294 | 6.62 |
ENSRNOT00000074109
|
Hist2h4a
|
histone cluster 2, H4 |
| chr6_-_131914028 | 6.20 |
ENSRNOT00000007602
|
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr3_+_114772603 | 6.13 |
ENSRNOT00000073569
|
MGC105649
|
hypothetical LOC302884 |
| chr2_+_208738132 | 5.98 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr19_-_29968424 | 5.86 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr17_+_10137974 | 5.82 |
ENSRNOT00000031935
|
Hk3
|
hexokinase 3 |
| chr1_+_88955135 | 5.81 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr5_+_163773509 | 5.73 |
ENSRNOT00000072167
|
LOC103692519
|
60S ribosomal protein L9 pseudogene |
| chr6_+_99282850 | 5.70 |
ENSRNOT00000008374
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 |
| chr16_+_8302950 | 5.65 |
ENSRNOT00000066062
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr16_+_75966352 | 5.36 |
ENSRNOT00000022774
|
Angpt2
|
angiopoietin 2 |
| chr1_-_222224910 | 5.34 |
ENSRNOT00000028720
|
Plcb3
|
phospholipase C beta 3 |
| chrX_-_15504165 | 5.30 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr12_+_47590154 | 5.06 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr8_-_119523964 | 5.01 |
ENSRNOT00000081718
|
Mlh1
|
mutL homolog 1 |
| chr1_+_88955440 | 4.97 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr6_-_115616766 | 4.94 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chrX_-_123486814 | 4.87 |
ENSRNOT00000016993
|
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr2_+_150013327 | 4.78 |
ENSRNOT00000075623
|
AABR07010747.1
|
|
| chr4_+_148782479 | 4.77 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr6_+_93461713 | 4.76 |
ENSRNOT00000031595
|
Arid4a
|
AT-rich interaction domain 4A |
| chr3_+_122662086 | 4.76 |
ENSRNOT00000009097
|
Tgm6
|
transglutaminase 6 |
| chrX_-_2116483 | 4.71 |
ENSRNOT00000055077
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr3_-_9936352 | 4.69 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr1_+_227743612 | 4.67 |
ENSRNOT00000028476
|
Gm8369
|
predicted gene 8369 |
| chr9_+_112360419 | 4.64 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr16_-_23991570 | 4.59 |
ENSRNOT00000044198
ENSRNOT00000018854 |
Nat2
Nat1
|
N-acetyltransferase 2 N-acetyltransferase 1 |
| chr12_+_51878153 | 4.43 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr20_+_8312792 | 4.36 |
ENSRNOT00000083108
|
Cmtr1
|
cap methyltransferase 1 |
| chr8_-_119523716 | 4.35 |
ENSRNOT00000064581
|
Mlh1
|
mutL homolog 1 |
| chr8_+_77107536 | 4.28 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr11_+_60140130 | 4.13 |
ENSRNOT00000033595
|
LOC685680
|
similar to TPA-induced transmembrane protein |
| chr20_-_10968432 | 4.09 |
ENSRNOT00000001593
|
Cstb
|
cystatin B |
| chr17_-_4454701 | 4.09 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr7_+_144865608 | 3.92 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr3_-_76518601 | 3.89 |
ENSRNOT00000071021
|
Olr623
|
olfactory receptor 623 |
| chr10_+_40812858 | 3.88 |
ENSRNOT00000018149
|
G3bp1
|
G3BP stress granule assembly factor 1 |
| chr20_+_29831314 | 3.87 |
ENSRNOT00000000696
|
Psap
|
prosaposin |
| chr5_+_58144705 | 3.79 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
| chr11_-_35749464 | 3.75 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr1_-_129776276 | 3.72 |
ENSRNOT00000051402
|
Arrdc4
|
arrestin domain containing 4 |
| chrX_-_2116656 | 3.69 |
ENSRNOT00000081913
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr3_+_160908769 | 3.65 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr1_+_48273611 | 3.64 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr19_+_24849356 | 3.45 |
ENSRNOT00000090312
|
Ddx39a
|
DExD-box helicase 39A |
| chr2_+_104854572 | 3.45 |
ENSRNOT00000064828
ENSRNOT00000090912 |
Hltf
|
helicase-like transcription factor |
| chr9_+_95202632 | 3.42 |
ENSRNOT00000025652
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_-_29949603 | 3.39 |
ENSRNOT00000009419
|
LOC103692829
|
60S ribosomal protein L9 pseudogene |
| chr1_+_260798239 | 3.39 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr5_-_73986373 | 3.29 |
ENSRNOT00000064043
ENSRNOT00000032299 |
Tmem245
|
transmembrane protein 245 |
| chr3_+_75322884 | 3.25 |
ENSRNOT00000066249
|
Olr556
|
olfactory receptor 556 |
| chr16_+_50179458 | 3.21 |
ENSRNOT00000041946
|
F11
|
coagulation factor XI |
| chr3_-_119405453 | 3.19 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr5_-_100970043 | 3.05 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr3_-_76346196 | 3.04 |
ENSRNOT00000047702
|
Olr613
|
olfactory receptor 613 |
| chr3_-_3890758 | 2.96 |
ENSRNOT00000064083
|
Sec16a
|
SEC16 homolog A, endoplasmic reticulum export factor |
| chr4_-_40136061 | 2.94 |
ENSRNOT00000009752
|
Bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr3_-_103203299 | 2.87 |
ENSRNOT00000047208
|
Olr783
|
olfactory receptor 783 |
| chr13_-_74520634 | 2.82 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_+_69960160 | 2.81 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chr5_-_155709215 | 2.76 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr20_+_44220840 | 2.75 |
ENSRNOT00000047194
|
Tube1
|
tubulin, epsilon 1 |
| chrX_+_112311251 | 2.73 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr5_-_56536772 | 2.65 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr17_-_32015071 | 2.60 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr5_+_121952977 | 2.58 |
ENSRNOT00000008278
|
Pde4b
|
phosphodiesterase 4B |
| chr3_-_75752501 | 2.56 |
ENSRNOT00000042725
|
Olr578
|
olfactory receptor 578 |
| chr8_-_77398156 | 2.54 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr11_+_81796891 | 2.53 |
ENSRNOT00000058402
|
Crygs
|
crystallin, gamma S |
| chrX_-_36884748 | 2.51 |
ENSRNOT00000065678
|
Phka2
|
phosphorylase kinase, alpha 2 |
| chr5_+_35991068 | 2.49 |
ENSRNOT00000061139
|
Pnisr
|
PNN interacting serine and arginine rich protein |
| chr9_-_52238564 | 2.44 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr6_+_8220228 | 2.32 |
ENSRNOT00000079279
ENSRNOT00000048656 |
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr1_+_168204985 | 2.31 |
ENSRNOT00000049036
|
Olr75
|
olfactory receptor 75 |
| chr2_+_208373154 | 2.27 |
ENSRNOT00000082943
ENSRNOT00000050538 |
LOC100911796
|
adenosine receptor A3-like |
| chr20_-_6864387 | 2.26 |
ENSRNOT00000068527
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr5_+_159734838 | 2.22 |
ENSRNOT00000079905
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chrX_+_1787266 | 2.20 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr13_-_91735361 | 2.18 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr2_+_222021103 | 2.16 |
ENSRNOT00000086125
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr6_+_49852130 | 2.15 |
ENSRNOT00000068208
|
Sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr17_+_57906050 | 2.12 |
ENSRNOT00000077338
|
AABR07028157.1
|
|
| chr13_+_27032048 | 2.12 |
ENSRNOT00000031789
|
Serpinb13
|
serpin family B member 13 |
| chr8_-_43970394 | 2.11 |
ENSRNOT00000079362
|
Olr1340
|
olfactory receptor 1340 |
| chr17_+_6909728 | 2.08 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr16_-_75208230 | 2.07 |
ENSRNOT00000058059
|
Defb10
|
defensin beta 10 |
| chr11_+_31893489 | 2.07 |
ENSRNOT00000042750
|
Itsn1
|
intersectin 1 |
| chr10_+_34489940 | 2.00 |
ENSRNOT00000085975
|
Zfp62
|
zinc finger protein 62 |
| chr2_+_243656579 | 1.98 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr3_+_75518280 | 1.97 |
ENSRNOT00000079476
|
Olr566
|
olfactory receptor 566 |
| chr10_-_98390384 | 1.86 |
ENSRNOT00000065947
|
Abca8a
|
ATP-binding cassette, subfamily A (ABC1), member 8a |
| chr15_-_70349983 | 1.80 |
ENSRNOT00000012167
|
Diaph3
|
diaphanous-related formin 3 |
| chr5_-_120743748 | 1.75 |
ENSRNOT00000057774
|
AABR07049401.1
|
|
| chr1_-_88428685 | 1.75 |
ENSRNOT00000074582
|
Zfp566
|
zinc finger protein 566 |
| chr9_-_14502933 | 1.74 |
ENSRNOT00000078464
|
Unc5cl
|
unc-5 family C-terminal like |
| chr17_-_39824299 | 1.73 |
ENSRNOT00000023412
|
Prl
|
prolactin |
| chr2_+_127525285 | 1.71 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr7_-_115979298 | 1.69 |
ENSRNOT00000075883
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr10_-_43998533 | 1.69 |
ENSRNOT00000047037
|
Olr1417
|
olfactory receptor 1417 |
| chr7_-_58365313 | 1.67 |
ENSRNOT00000005443
|
Thap2
|
THAP domain containing 2 |
| chr14_-_46054022 | 1.67 |
ENSRNOT00000002982
|
LOC498368
|
similar to RIKEN cDNA 0610040J01 |
| chr2_+_226563050 | 1.59 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_13892997 | 1.59 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr3_+_75809445 | 1.56 |
ENSRNOT00000049950
|
Olr581
|
olfactory receptor 581 |
| chr3_+_158887412 | 1.49 |
ENSRNOT00000075772
|
Olr1261
|
olfactory receptor 1261 |
| chr1_+_261291870 | 1.46 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chrX_+_44907521 | 1.45 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr16_+_18716019 | 1.43 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr20_+_27173213 | 1.41 |
ENSRNOT00000073617
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr20_+_47395014 | 1.37 |
ENSRNOT00000057116
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr3_-_77402579 | 1.37 |
ENSRNOT00000043801
|
Olr658
|
olfactory receptor 658 |
| chr10_-_62361963 | 1.33 |
ENSRNOT00000072702
|
LOC108348032
|
olfactory receptor 1 |
| chr5_-_69171311 | 1.33 |
ENSRNOT00000072957
|
LOC100912327
|
olfactory receptor 13C3-like |
| chr3_-_431933 | 1.31 |
ENSRNOT00000033653
|
Spopl
|
speckle type BTB/POZ protein like |
| chr7_+_6215743 | 1.31 |
ENSRNOT00000050967
|
Olr1002
|
olfactory receptor 1002 |
| chr3_-_146396299 | 1.26 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr3_+_154437571 | 1.26 |
ENSRNOT00000019331
|
AABR07054456.1
|
|
| chrX_-_105417323 | 1.21 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr1_-_55895636 | 1.21 |
ENSRNOT00000029537
|
Vom2r9
|
vomeronasal 2 receptor, 9 |
| chr3_+_20965751 | 1.21 |
ENSRNOT00000010450
|
Olr417
|
olfactory receptor 417 |
| chr12_-_38916237 | 1.16 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr10_-_44659707 | 1.14 |
ENSRNOT00000002064
|
RGD1559534
|
similar to Alpha enolase (2-phospho-D-glycerate hydro-lyase) |
| chr1_-_7443863 | 1.12 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr2_+_165805144 | 1.11 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr2_+_257568613 | 1.11 |
ENSRNOT00000066125
|
Usp33
|
ubiquitin specific peptidase 33 |
| chr11_-_33801999 | 1.08 |
ENSRNOT00000002308
|
Setd4
|
SET domain containing 4 |
| chr1_+_172860797 | 1.07 |
ENSRNOT00000081313
|
LOC687082
|
similar to olfactory receptor 508 |
| chr18_+_3887419 | 1.05 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr10_-_44026578 | 1.05 |
ENSRNOT00000050448
|
Olr1418
|
olfactory receptor 1418 |
| chr2_+_208323882 | 1.04 |
ENSRNOT00000085178
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr3_-_71012860 | 1.03 |
ENSRNOT00000078440
|
AABR07052729.1
|
|
| chrM_+_11736 | 1.02 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr20_+_572454 | 1.01 |
ENSRNOT00000061839
|
Olr1684
|
olfactory receptor 1684 |
| chr17_-_63879166 | 1.01 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr11_-_67551211 | 0.99 |
ENSRNOT00000049463
|
Stfa3l1
|
stefin A3-like 1 |
| chr6_+_11644578 | 0.99 |
ENSRNOT00000021923
ENSRNOT00000093689 |
Msh6
|
mutS homolog 6 |
| chr6_+_136380990 | 0.98 |
ENSRNOT00000063865
ENSRNOT00000082914 |
Zfyve21
|
zinc finger FYVE-type containing 21 |
| chr2_+_243018975 | 0.97 |
ENSRNOT00000081989
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr6_+_76079880 | 0.97 |
ENSRNOT00000009304
|
RGD1305089
|
similar to 1110008L16Rik protein |
| chr4_-_66062089 | 0.95 |
ENSRNOT00000018782
|
Zc3hav1
|
zinc finger CCCH-type containing, antiviral 1 |
| chr15_-_18521161 | 0.94 |
ENSRNOT00000051556
|
LOC498460
|
LRRGT00055 |
| chr5_-_100977902 | 0.93 |
ENSRNOT00000077429
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr1_-_169032157 | 0.82 |
ENSRNOT00000034624
|
Olr129
|
olfactory receptor 129 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 6.7 | 20.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 6.1 | 18.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 5.4 | 16.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 4.8 | 28.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 3.9 | 11.7 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 3.6 | 10.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 3.1 | 9.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 3.1 | 21.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 2.8 | 52.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 2.7 | 13.4 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 2.6 | 13.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 2.0 | 8.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.9 | 5.7 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.9 | 15.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.8 | 24.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.8 | 5.3 | GO:1905076 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 1.5 | 4.4 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 1.4 | 4.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.3 | 13.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.3 | 5.3 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 1.3 | 18.9 | GO:0051764 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 1.3 | 3.8 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 1.2 | 9.3 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 1.1 | 8.6 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 1.0 | 7.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.0 | 6.2 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 1.0 | 9.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 1.0 | 3.9 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.9 | 8.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.9 | 2.8 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.8 | 2.5 | GO:0010034 | response to acetate(GO:0010034) |
| 0.8 | 16.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.8 | 2.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.2 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.7 | 2.2 | GO:0046127 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.7 | 2.8 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.7 | 4.8 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.7 | 2.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.7 | 2.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.6 | 3.9 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.6 | 4.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.6 | 3.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.6 | 20.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.5 | 4.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 5.8 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.5 | 16.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.5 | 3.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 2.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 2.1 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.4 | 4.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 6.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.4 | 9.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.4 | 43.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.4 | 4.6 | GO:0097068 | response to thyroxine(GO:0097068) |
| 0.4 | 3.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 3.6 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.3 | 9.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 3.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 5.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 3.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 5.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 3.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.2 | 4.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 2.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 1.0 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.2 | 4.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 1.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 2.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 10.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 2.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 5.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 9.6 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.1 | 3.7 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 13.0 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.1 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 19.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 4.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.4 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.1 | 2.1 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 2.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 3.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 3.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 2.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.7 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 4.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 3.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 6.3 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 2.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 1.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 1.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 1.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 4.7 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0005713 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 2.9 | 11.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 2.8 | 52.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.6 | 4.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 1.5 | 7.3 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 1.3 | 3.9 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.9 | 8.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.7 | 11.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.7 | 18.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 2.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 4.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 13.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.5 | 7.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 1.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.5 | 18.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 2.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 3.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 13.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.4 | 6.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 9.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 5.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.4 | 10.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 2.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 22.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 88.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 9.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 15.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 6.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 15.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 1.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 6.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 8.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 10.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 4.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.1 | 4.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.4 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 3.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 3.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 12.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 8.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 52.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.6 | 10.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 3.0 | 21.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 3.0 | 11.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.7 | 21.8 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 2.3 | 11.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 2.2 | 20.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 2.1 | 10.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 2.0 | 5.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.5 | 4.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.5 | 4.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.4 | 5.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.3 | 13.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.3 | 10.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 1.2 | 3.6 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.0 | 10.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.0 | 5.8 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.9 | 2.8 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.9 | 5.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 16.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.8 | 3.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.7 | 2.9 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.7 | 2.2 | GO:0017113 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.7 | 2.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.6 | 7.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.6 | 3.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.5 | 3.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 13.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 5.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.5 | 4.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 20.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 20.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 2.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 1.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 2.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 4.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 2.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 0.8 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.3 | 3.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 5.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 18.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 75.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 6.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 4.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 11.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 11.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 4.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 9.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 24.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 15.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 4.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 9.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 46.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0034235 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 0.1 | 2.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 7.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 6.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 2.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 3.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 9.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 17.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 4.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 11.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 5.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 4.8 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 45.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.9 | 50.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.7 | 13.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 11.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 15.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 18.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 13.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.3 | 5.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.3 | 2.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 7.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 11.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 40.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 25.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 9.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 7.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 5.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 20.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.1 | 13.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.1 | 11.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.9 | 18.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.9 | 13.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.8 | 9.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.8 | 24.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.7 | 11.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.6 | 21.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.6 | 9.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 13.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.5 | 5.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 11.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 4.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 3.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 5.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 4.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 6.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 2.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 7.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 4.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 4.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 5.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 7.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 16.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.2 | 3.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 10.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 4.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 5.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 10.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 2.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 3.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |