Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
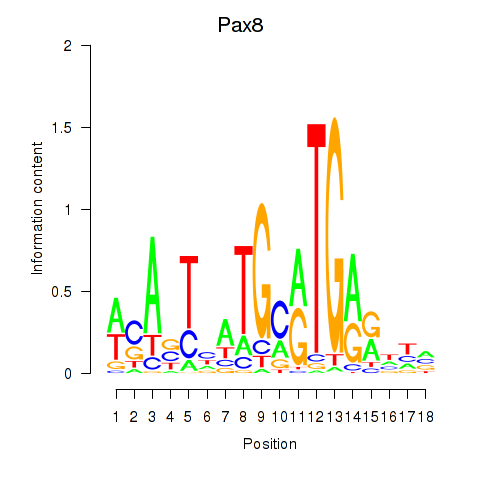
Results for Pax8
Z-value: 0.28
Transcription factors associated with Pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax8
|
ENSRNOG00000026203 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax8 | rn6_v1_chr3_-_1549189_1549189 | -0.33 | 8.5e-10 | Click! |
Activity profile of Pax8 motif
Sorted Z-values of Pax8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89498395 | 9.04 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr8_-_111721303 | 7.51 |
ENSRNOT00000045628
ENSRNOT00000012725 |
Tf
|
transferrin |
| chr8_+_36125999 | 6.99 |
ENSRNOT00000013036
ENSRNOT00000088046 |
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chr2_-_231409496 | 6.46 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr8_-_116893057 | 6.41 |
ENSRNOT00000082113
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr18_-_62614725 | 6.29 |
ENSRNOT00000025223
|
Mc4r
|
melanocortin 4 receptor |
| chr7_+_28654733 | 5.66 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_-_231409988 | 5.62 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr17_+_76532611 | 5.14 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_-_220878059 | 4.70 |
ENSRNOT00000036745
|
Snx32
|
sorting nexin 32 |
| chr6_-_23543172 | 4.24 |
ENSRNOT00000006073
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr4_-_119591817 | 3.94 |
ENSRNOT00000048768
|
AC097129.1
|
|
| chr6_-_23542928 | 3.87 |
ENSRNOT00000083858
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr4_-_144869919 | 3.41 |
ENSRNOT00000009066
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr5_+_19284548 | 3.40 |
ENSRNOT00000012551
|
Ubxn2b
|
UBX domain protein 2B |
| chr4_+_149908375 | 3.23 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr7_+_93975451 | 2.99 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr15_+_30926938 | 2.57 |
ENSRNOT00000075625
|
AABR07017773.1
|
|
| chr10_-_56409017 | 2.41 |
ENSRNOT00000020152
|
Fgf11
|
fibroblast growth factor 11 |
| chr15_-_29623597 | 1.95 |
ENSRNOT00000074483
|
AABR07017647.1
|
|
| chr4_-_56453563 | 1.94 |
ENSRNOT00000060362
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr8_-_114010277 | 1.83 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr1_-_87777359 | 1.55 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chr14_-_11546314 | 1.49 |
ENSRNOT00000090334
|
AABR07014355.1
|
|
| chr10_-_93679974 | 1.42 |
ENSRNOT00000009316
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr1_-_62316450 | 1.39 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr5_+_77052484 | 1.38 |
ENSRNOT00000059419
ENSRNOT00000087254 |
Snx30
|
sorting nexin family member 30 |
| chr1_-_227047290 | 1.38 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chr20_-_27208041 | 1.33 |
ENSRNOT00000084720
|
Rufy2
|
RUN and FYVE domain containing 2 |
| chr17_+_15194262 | 1.25 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chrX_+_20216587 | 1.16 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr9_+_81868265 | 0.74 |
ENSRNOT00000022632
|
Bcs1l
|
BCS1 homolog, ubiquinol-cytochrome c reductase complex chaperone |
| chr17_+_14516566 | 0.72 |
ENSRNOT00000060660
ENSRNOT00000071469 |
LOC689569
|
similar to vitamin A-deficient testicular protein 11-like |
| chr1_+_80973739 | 0.70 |
ENSRNOT00000026088
ENSRNOT00000081983 |
AC118165.1
|
|
| chr7_-_15821927 | 0.60 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr17_-_36438752 | 0.58 |
ENSRNOT00000048997
|
RGD1561310
|
similar to ribosomal protein L37 |
| chr7_+_8210182 | 0.58 |
ENSRNOT00000075032
|
Olr1049
|
olfactory receptor 1049 |
| chr15_+_30910039 | 0.39 |
ENSRNOT00000072686
|
AABR07017772.1
|
|
| chr9_-_81868086 | 0.39 |
ENSRNOT00000067080
|
Zfp142
|
zinc finger protein 142 |
| chr16_-_54628458 | 0.39 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr1_-_230079916 | 0.38 |
ENSRNOT00000091024
|
Olr358
|
olfactory receptor 358 |
| chr14_+_22517774 | 0.37 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_102826965 | 0.35 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr2_-_210473761 | 0.29 |
ENSRNOT00000066501
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr4_-_87982729 | 0.21 |
ENSRNOT00000089498
|
Vom1r79
|
vomeronasal 1 receptor 79 |
| chr13_-_34253910 | 0.19 |
ENSRNOT00000092847
|
Tsn
|
translin |
| chr13_-_111948753 | 0.14 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr17_+_26772096 | 0.09 |
ENSRNOT00000071866
|
AABR07027368.1
|
|
| chr4_-_150829741 | 0.01 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.9 | 7.5 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.7 | 7.0 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 1.1 | 5.7 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.1 | 6.4 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.9 | 5.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 6.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.6 | 1.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.2 | 3.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 8.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.4 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.7 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 6.1 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 3.4 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.3 | 7.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 12.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 7.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 6.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.9 | 7.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.9 | 6.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 12.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 5.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 5.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 3.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 6.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 7.5 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 6.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 7.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 8.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |