Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
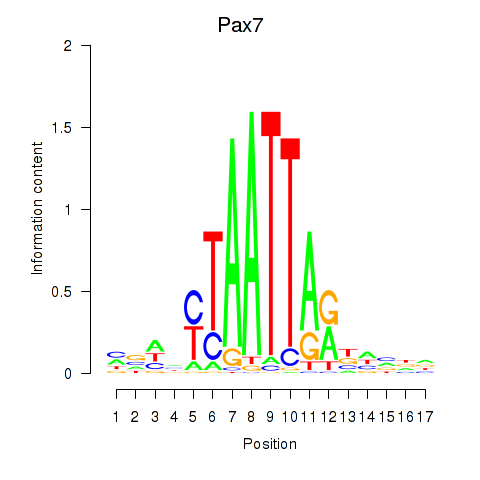
Results for Pax7
Z-value: 0.48
Transcription factors associated with Pax7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax7
|
ENSRNOG00000018739 | paired box 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax7 | rn6_v1_chr5_-_158313426_158313426 | -0.20 | 3.5e-04 | Click! |
Activity profile of Pax7 motif
Sorted Z-values of Pax7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_5709236 | 18.28 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr17_+_43423111 | 18.24 |
ENSRNOT00000022630
|
Hist1h2ba
|
histone cluster 1 H2B family member a |
| chr10_-_103826448 | 16.85 |
ENSRNOT00000085636
|
Fdxr
|
ferredoxin reductase |
| chr1_-_227457629 | 14.61 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr9_+_53013413 | 14.35 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr7_-_28711761 | 8.65 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr3_+_70327193 | 7.94 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr1_+_201337416 | 7.15 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr2_+_104020955 | 6.95 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr10_-_74679858 | 6.81 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr5_+_28485619 | 6.50 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr8_-_6305033 | 6.22 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr11_-_61234944 | 6.00 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr18_+_45023932 | 5.98 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr14_+_23405717 | 5.80 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr11_-_43022565 | 5.62 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr20_+_32717564 | 5.46 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr16_+_9023387 | 5.07 |
ENSRNOT00000067761
|
RGD1561145
|
similar to novel protein |
| chr18_-_13183263 | 4.95 |
ENSRNOT00000050933
|
Ccdc178
|
coiled-coil domain containing 178 |
| chr2_+_202200797 | 4.87 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr1_+_128637049 | 4.83 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr3_-_8659102 | 4.80 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chrX_+_14019961 | 4.72 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr7_+_78558701 | 4.54 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_183424449 | 4.49 |
ENSRNOT00000071930
|
Fam60a
|
family with sequence similarity 60, member A |
| chr6_+_76349362 | 4.47 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr18_+_79773608 | 4.39 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr4_-_80395502 | 4.32 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr10_-_65502936 | 4.29 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr1_+_101603222 | 4.25 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr1_+_15834779 | 4.15 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr6_-_108660063 | 4.13 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr3_-_165700489 | 3.82 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr9_+_8052210 | 3.77 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr2_-_33025271 | 3.62 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_224998172 | 3.61 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr15_+_31950986 | 3.57 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr9_+_60039297 | 3.48 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr13_-_76049363 | 3.44 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr16_-_29936307 | 3.35 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr9_+_71915421 | 3.34 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chrX_+_131381134 | 3.31 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr2_+_80269661 | 3.22 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr1_-_261179790 | 3.15 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr17_-_30661690 | 3.09 |
ENSRNOT00000048146
|
RGD1307537
|
similar to RIKEN cDNA 4933417A18 |
| chr3_+_75906945 | 3.05 |
ENSRNOT00000047110
|
Olr586
|
olfactory receptor 586 |
| chr8_+_104106740 | 2.94 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr9_-_78607307 | 2.71 |
ENSRNOT00000057672
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr18_+_28594892 | 2.60 |
ENSRNOT00000067702
|
Ube2d2
|
ubiquitin-conjugating enzyme E2D 2 |
| chrX_+_105147534 | 2.56 |
ENSRNOT00000046288
|
Cenpi
|
centromere protein I |
| chr2_+_66940057 | 2.54 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr1_+_101554642 | 2.54 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr3_-_90751055 | 2.49 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr7_+_2459141 | 2.27 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chrX_+_105147153 | 2.10 |
ENSRNOT00000088172
|
Cenpi
|
centromere protein I |
| chr4_+_147832136 | 2.01 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr2_+_198417619 | 1.99 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr2_-_122646445 | 1.97 |
ENSRNOT00000080100
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr5_-_162751128 | 1.97 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chr7_-_69982592 | 1.82 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr3_+_3389612 | 1.74 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr10_+_29289203 | 1.61 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr7_-_64887448 | 1.50 |
ENSRNOT00000038147
|
Helb
|
DNA helicase B |
| chr12_-_45801842 | 1.49 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr2_-_170301348 | 1.48 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr7_-_121000272 | 1.43 |
ENSRNOT00000020941
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr10_-_57837602 | 1.41 |
ENSRNOT00000075185
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr3_+_138398011 | 1.41 |
ENSRNOT00000038865
|
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr7_+_2752680 | 1.40 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr3_+_73327828 | 1.31 |
ENSRNOT00000071299
|
LOC684683
|
similar to olfactory receptor Olr470 |
| chr10_-_88670430 | 1.28 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chrX_-_40086870 | 1.08 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr17_-_78735324 | 0.93 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr16_+_48513432 | 0.84 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr4_+_156253079 | 0.75 |
ENSRNOT00000013536
|
Clec4d
|
C-type lectin domain family 4, member D |
| chr4_+_82637411 | 0.68 |
ENSRNOT00000048225
|
LOC685406
|
LRRGT00062 |
| chr6_+_145546595 | 0.65 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr8_+_43781921 | 0.63 |
ENSRNOT00000078003
|
Olr1329
|
olfactory receptor 1329 |
| chr18_-_15688284 | 0.61 |
ENSRNOT00000091816
|
Dsg3
|
desmoglein 3 |
| chr2_-_58534211 | 0.59 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chrM_+_7006 | 0.56 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr10_-_67401836 | 0.51 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr2_+_127525285 | 0.51 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr4_-_165832323 | 0.48 |
ENSRNOT00000007497
|
Tas2r114
|
taste receptor, type 2, member 114 |
| chr10_-_87521514 | 0.43 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr10_+_61640015 | 0.41 |
ENSRNOT00000092714
|
Mettl16
|
methyltransferase like 16 |
| chr8_+_5790034 | 0.39 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr14_-_16903242 | 0.28 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr3_+_73366155 | 0.28 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
| chr17_+_78735598 | 0.28 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr12_+_47590154 | 0.23 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr9_+_23596964 | 0.23 |
ENSRNOT00000064279
|
LOC108351994
|
exocrine gland-secreted peptide 1-like |
| chr4_-_88684415 | 0.05 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 3.0 | 18.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.7 | 16.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.2 | 4.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.0 | 10.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.8 | 2.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.8 | 2.3 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.7 | 4.3 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.6 | 5.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.5 | 5.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.5 | 2.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 5.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 4.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 6.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 4.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.4 | 3.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 4.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 4.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 6.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 6.0 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 2.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 1.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 8.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 3.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 4.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 4.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 5.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 2.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 4.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 1.5 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 2.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 4.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.4 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 2.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 3.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 4.2 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.7 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.5 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 1.0 | 4.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.5 | 2.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 4.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 3.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 3.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 4.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 20.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 6.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 1.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 4.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 2.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 18.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 12.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 4.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 8.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 8.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 12.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.6 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.4 | 4.3 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 1.1 | 5.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.9 | 16.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.8 | 2.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.6 | 4.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 6.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.5 | 1.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.5 | 4.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 2.7 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.4 | 3.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 1.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 2.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 1.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 4.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 3.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 2.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 4.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 4.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 24.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 5.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 5.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 18.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 16.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 18.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 3.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 2.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 1.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 4.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |