Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
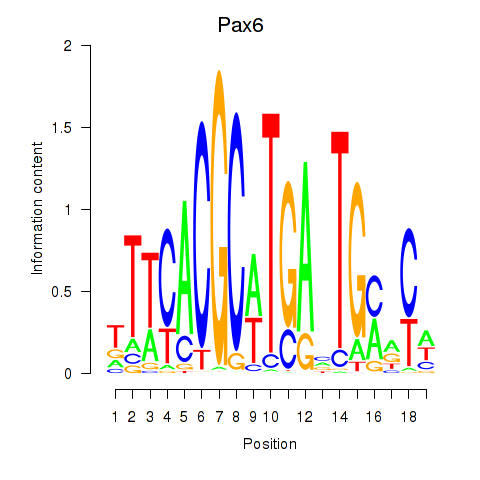
Results for Pax6
Z-value: 0.72
Transcription factors associated with Pax6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax6
|
ENSRNOG00000004410 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax6 | rn6_v1_chr3_+_95711555_95711555 | 0.06 | 2.8e-01 | Click! |
Activity profile of Pax6 motif
Sorted Z-values of Pax6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_197821936 | 24.68 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr4_+_98457810 | 24.41 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr3_+_16413080 | 21.59 |
ENSRNOT00000040386
|
LOC100912707
|
Ig kappa chain V19-17-like |
| chr8_-_49280901 | 17.78 |
ENSRNOT00000021390
|
Cd3g
|
CD3g molecule |
| chr17_-_44840131 | 17.67 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr6_-_143195445 | 15.59 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr4_-_102124609 | 14.90 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr2_+_189997129 | 14.75 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chr15_+_62406873 | 14.33 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr6_-_140216072 | 14.31 |
ENSRNOT00000072365
|
AABR07065750.1
|
|
| chr3_-_52849907 | 13.17 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr3_+_16846412 | 13.12 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr6_-_143195143 | 12.83 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr9_-_103207190 | 12.51 |
ENSRNOT00000026010
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr1_-_280015358 | 12.11 |
ENSRNOT00000024548
|
Hspa12a
|
heat shock protein family A (Hsp70) member 12A |
| chr15_+_32614002 | 11.99 |
ENSRNOT00000072962
|
AABR07017902.1
|
|
| chr1_-_141470380 | 11.80 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chr15_-_28081465 | 11.77 |
ENSRNOT00000033739
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr3_+_45538797 | 11.70 |
ENSRNOT00000007632
|
Dapl1
|
death associated protein-like 1 |
| chr6_-_140215907 | 10.61 |
ENSRNOT00000086370
|
AABR07065750.1
|
|
| chr11_-_84931160 | 10.61 |
ENSRNOT00000071848
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr18_+_55666027 | 10.51 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chrX_-_77295426 | 10.34 |
ENSRNOT00000090833
|
Taf9b
|
TATA-box binding protein associated factor 9b |
| chr18_+_55463308 | 10.14 |
ENSRNOT00000073388
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr14_+_71542057 | 10.06 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr2_-_88553086 | 9.38 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chrX_+_159158194 | 9.01 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr7_+_42304534 | 8.77 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr17_+_44738643 | 8.76 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr6_-_102196138 | 8.75 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr1_+_86938138 | 8.69 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr13_+_92264231 | 8.56 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr1_-_23556241 | 8.29 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr1_-_225952516 | 8.22 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr10_+_59652549 | 7.98 |
ENSRNOT00000073209
|
Itgae
|
integrin subunit alpha E |
| chr4_+_102068556 | 7.91 |
ENSRNOT00000077412
|
AABR07060971.1
|
|
| chr4_-_179307095 | 7.79 |
ENSRNOT00000021193
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr2_+_187322416 | 7.52 |
ENSRNOT00000025183
|
Crabp2
|
cellular retinoic acid binding protein 2 |
| chr5_+_134691881 | 7.44 |
ENSRNOT00000091543
ENSRNOT00000067655 |
Mknk1
|
MAP kinase-interacting serine/threonine kinase 1 |
| chr19_+_10563423 | 7.37 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr20_-_45259928 | 7.16 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chrX_+_120859968 | 7.06 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr8_-_130429132 | 6.93 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr7_-_70476340 | 6.89 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chr1_-_80271001 | 6.65 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr18_+_402295 | 6.65 |
ENSRNOT00000033618
|
Fundc2
|
FUN14 domain containing 2 |
| chr6_-_55647665 | 6.63 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_30869628 | 6.36 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr10_+_11046024 | 6.29 |
ENSRNOT00000084850
|
Nmral1
|
NmrA like redox sensor 1 |
| chr3_+_17009089 | 6.26 |
ENSRNOT00000048829
|
LOC100361052
|
rCG64257-like |
| chr12_-_2661565 | 6.24 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr12_+_37709860 | 6.20 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chrX_+_120860178 | 6.13 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr2_-_41785792 | 6.13 |
ENSRNOT00000015871
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr5_+_141491223 | 6.02 |
ENSRNOT00000034839
|
Rhbdl2
|
rhomboid like 2 |
| chr12_-_22472044 | 5.89 |
ENSRNOT00000073356
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr9_-_11108741 | 5.85 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr4_+_101949285 | 5.82 |
ENSRNOT00000058446
|
AABR07060963.3
|
|
| chr11_+_43329700 | 5.66 |
ENSRNOT00000060892
|
Olr1540
|
olfactory receptor 1540 |
| chr20_-_30327361 | 5.65 |
ENSRNOT00000000689
|
Slc29a3
|
solute carrier family 29 member 3 |
| chr6_-_26546545 | 5.61 |
ENSRNOT00000091264
|
Snx17
|
sorting nexin 17 |
| chr1_-_166943592 | 5.60 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr12_-_13157570 | 5.57 |
ENSRNOT00000001437
ENSRNOT00000067332 ENSRNOT00000092238 |
Daglb
|
diacylglycerol lipase, beta |
| chr3_-_12944494 | 5.54 |
ENSRNOT00000023172
|
Mvb12b
|
multivesicular body subunit 12B |
| chr13_-_113872097 | 5.51 |
ENSRNOT00000010799
ENSRNOT00000084320 |
Cr1l
|
complement C3b/C4b receptor 1 like |
| chr18_+_30880020 | 5.49 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr5_-_148392689 | 5.46 |
ENSRNOT00000018464
ENSRNOT00000080166 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr2_+_116032455 | 5.40 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr14_-_33164141 | 5.38 |
ENSRNOT00000002844
|
Rest
|
RE1-silencing transcription factor |
| chr10_-_25890639 | 5.36 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr12_-_36555694 | 5.33 |
ENSRNOT00000001292
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr2_-_205212681 | 5.28 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr16_+_26906716 | 5.27 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr20_-_7142213 | 5.23 |
ENSRNOT00000078918
|
RGD1564450
|
RGD1564450 |
| chr20_-_45260119 | 5.17 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr20_+_4369178 | 5.16 |
ENSRNOT00000088079
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr15_-_27815261 | 5.09 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr10_+_109073678 | 4.97 |
ENSRNOT00000005379
|
Chmp6
|
charged multivesicular body protein 6 |
| chr10_+_58995556 | 4.81 |
ENSRNOT00000045100
|
Ggt6
|
gamma-glutamyl transferase 6 |
| chr2_+_87418517 | 4.77 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr18_+_30808404 | 4.67 |
ENSRNOT00000060475
|
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr1_-_31528083 | 4.66 |
ENSRNOT00000061041
|
Rpl26-ps2
|
ribosomal protein L26, pseudogene 2 |
| chr4_+_14212925 | 4.62 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr5_+_64892802 | 4.61 |
ENSRNOT00000059869
|
Rnf20
|
ring finger protein 20 |
| chr12_+_2948570 | 4.58 |
ENSRNOT00000071450
|
LOC100363914
|
hypothetical LOC100363914 |
| chr1_+_114453275 | 4.53 |
ENSRNOT00000019245
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr5_+_153260930 | 4.49 |
ENSRNOT00000023289
ENSRNOT00000082184 |
Rsrp1
|
arginine and serine rich protein 1 |
| chr1_+_207654487 | 4.47 |
ENSRNOT00000025569
|
Foxi2
|
forkhead box I2 |
| chr10_-_65424802 | 4.46 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr3_+_117422704 | 4.33 |
ENSRNOT00000085647
|
Slc12a1
|
solute carrier family 12 member 1 |
| chr18_+_63023930 | 4.19 |
ENSRNOT00000085655
|
Impa2
|
inositol monophosphatase 2 |
| chr10_+_53713938 | 4.16 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr2_+_188139730 | 4.06 |
ENSRNOT00000056789
ENSRNOT00000077698 |
Gon4l
|
gon-4 like |
| chr7_+_13318533 | 3.83 |
ENSRNOT00000034545
|
LOC691170
|
similar to zinc finger protein 84 (HPF2) |
| chr3_+_159995064 | 3.83 |
ENSRNOT00000012606
|
Ttpal
|
alpha tocopherol transfer protein like |
| chr20_+_1322453 | 3.82 |
ENSRNOT00000075472
|
Olr1868
|
olfactory receptor 1868 |
| chr1_-_166911694 | 3.78 |
ENSRNOT00000066915
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr10_-_58771908 | 3.74 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr5_-_141430659 | 3.62 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr10_+_46940965 | 3.51 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr14_+_86029335 | 3.35 |
ENSRNOT00000017375
|
Dbnl
|
drebrin-like |
| chr11_-_77703255 | 3.27 |
ENSRNOT00000083319
|
Cldn16
|
claudin 16 |
| chr20_+_3230052 | 3.16 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr8_-_49502647 | 3.14 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr15_+_32386816 | 3.14 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chrX_-_15707436 | 3.07 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr3_+_128828331 | 2.96 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr1_+_254697247 | 2.96 |
ENSRNOT00000025412
|
Rpp30
|
ribonuclease P/MRP subunit p30 |
| chr1_-_87777668 | 2.95 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr17_-_2705123 | 2.85 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr3_-_96065711 | 2.77 |
ENSRNOT00000006431
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr14_-_21878989 | 2.70 |
ENSRNOT00000072327
|
Dzip1-ps1
|
DAZ interacting protein 1, pseudogene 1 |
| chr10_-_25847994 | 2.67 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr2_+_166140112 | 2.60 |
ENSRNOT00000074564
|
Ppm1l
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr20_+_4369394 | 2.55 |
ENSRNOT00000088734
ENSRNOT00000091491 |
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr13_+_48679774 | 2.53 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr1_+_214215992 | 2.48 |
ENSRNOT00000051755
ENSRNOT00000023376 |
Phrf1
|
PHD and ring finger domains 1 |
| chr1_+_61268248 | 2.40 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr1_-_87777359 | 2.30 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chr4_-_119399130 | 2.28 |
ENSRNOT00000042796
|
Vom1r101
|
vomeronasal 1 receptor 101 |
| chr1_-_4653210 | 2.25 |
ENSRNOT00000019121
|
Rab32
|
RAB32, member RAS oncogene family |
| chr9_+_52687868 | 2.23 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr1_-_154170409 | 2.16 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr12_+_38465130 | 2.15 |
ENSRNOT00000080785
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr5_+_151573092 | 2.13 |
ENSRNOT00000011049
|
Slc9a1
|
solute carrier family 9 member A1 |
| chr6_-_8344897 | 2.12 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr15_+_18941431 | 2.11 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr16_-_19583386 | 1.92 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chrX_+_23146085 | 1.88 |
ENSRNOT00000068703
|
Apex2
|
apurinic/apyrimidinic endodeoxyribonuclease 2 |
| chr1_+_168575090 | 1.83 |
ENSRNOT00000048299
|
Olr103
|
olfactory receptor 103 |
| chrX_-_63291107 | 1.74 |
ENSRNOT00000092019
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr3_+_121632043 | 1.73 |
ENSRNOT00000024882
|
Polr1b
|
RNA polymerase I subunit B |
| chr2_+_78245459 | 1.71 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr11_+_87549059 | 1.70 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr11_+_76147205 | 1.68 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr3_-_12467662 | 1.58 |
ENSRNOT00000051893
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr1_+_1771710 | 1.54 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chr3_+_102619672 | 1.54 |
ENSRNOT00000040285
|
Olr756
|
olfactory receptor 756 |
| chr1_+_72580424 | 1.46 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr9_+_20004280 | 1.42 |
ENSRNOT00000075670
|
Ankrd66
|
ankyrin repeat domain 66 |
| chrX_+_136476823 | 1.36 |
ENSRNOT00000093183
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr15_+_30841153 | 1.31 |
ENSRNOT00000088305
|
LOC100911282
|
uncharacterized LOC100911282 |
| chr16_+_20704807 | 1.28 |
ENSRNOT00000027241
|
Klhl26
|
kelch-like family member 26 |
| chr20_-_29558321 | 1.25 |
ENSRNOT00000000702
|
Anapc16
|
anaphase promoting complex subunit 16 |
| chr1_-_79690434 | 1.22 |
ENSRNOT00000057986
ENSRNOT00000057988 |
LOC102557319
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr11_+_43161181 | 1.22 |
ENSRNOT00000071470
|
Olr1530
|
olfactory receptor 1530 |
| chr15_+_93612971 | 1.17 |
ENSRNOT00000013182
|
Acod1
|
aconitate decarboxylase 1 |
| chr5_+_134679713 | 1.16 |
ENSRNOT00000067566
|
Mob3c
|
MOB kinase activator 3C |
| chr10_-_54967585 | 1.16 |
ENSRNOT00000005255
|
Ntn1
|
netrin 1 |
| chr5_-_2982603 | 1.09 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr9_-_92963697 | 1.06 |
ENSRNOT00000023546
|
Gpr55
|
G protein-coupled receptor 55 |
| chr10_-_39054142 | 0.97 |
ENSRNOT00000063772
|
Rad50
|
RAD50 double strand break repair protein |
| chr4_-_33761163 | 0.97 |
ENSRNOT00000010079
|
Asns
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr2_-_198834038 | 0.94 |
ENSRNOT00000031484
|
Nudt17
|
nudix hydrolase 17 |
| chr2_+_205525204 | 0.94 |
ENSRNOT00000091210
|
Csde1
|
cold shock domain containing E1 |
| chr7_-_115963046 | 0.92 |
ENSRNOT00000007923
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr5_+_103479767 | 0.86 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr3_-_66279155 | 0.76 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr9_-_24451435 | 0.74 |
ENSRNOT00000060805
|
Defb49
|
defensin beta 49 |
| chr4_-_66955732 | 0.64 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr5_-_59208058 | 0.64 |
ENSRNOT00000020943
|
Olr834
|
olfactory receptor 834 |
| chr1_-_84812486 | 0.58 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr4_-_64981384 | 0.55 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr19_+_11450760 | 0.55 |
ENSRNOT00000026297
|
Nudt21
|
nudix hydrolase 21 |
| chr6_+_97110158 | 0.51 |
ENSRNOT00000088430
|
Syt16
|
synaptotagmin 16 |
| chr8_-_50126413 | 0.45 |
ENSRNOT00000072712
ENSRNOT00000022583 |
Cep164
|
centrosomal protein 164 |
| chr3_+_102553317 | 0.37 |
ENSRNOT00000043765
|
Olr753
|
olfactory receptor 753 |
| chr8_-_117237229 | 0.36 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr17_-_69862110 | 0.36 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr4_+_87608301 | 0.35 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr1_+_149963547 | 0.35 |
ENSRNOT00000042245
|
Olr20
|
olfactory receptor 20 |
| chr1_-_230729954 | 0.34 |
ENSRNOT00000017631
|
Olr382
|
olfactory receptor 382 |
| chr16_-_10952549 | 0.28 |
ENSRNOT00000091315
|
Opn4
|
opsin 4 |
| chr14_-_85982794 | 0.25 |
ENSRNOT00000076874
|
Urgcp
|
upregulator of cell proliferation |
| chr12_+_7208850 | 0.23 |
ENSRNOT00000001219
|
Katnal1
|
katanin catalytic subunit A1 like 1 |
| chr14_+_36071376 | 0.22 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr1_-_62316450 | 0.17 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr11_-_84090259 | 0.16 |
ENSRNOT00000002321
|
Eif2b5
|
eukaryotic translation initiation factor 2B subunit 5 epsilon |
| chr5_+_173152964 | 0.14 |
ENSRNOT00000024349
|
Ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr6_-_1466201 | 0.08 |
ENSRNOT00000089185
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr12_+_4546287 | 0.06 |
ENSRNOT00000001416
|
Higd2al1
|
HIG1 hypoxia inducible domain family, member 2A-like 1 |
| chr7_+_80750725 | 0.03 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr3_-_73854363 | 0.01 |
ENSRNOT00000051169
|
Olr496
|
olfactory receptor 496 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 2.5 | 10.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 2.5 | 12.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 2.5 | 12.3 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 1.9 | 5.6 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 1.8 | 24.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.8 | 8.8 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.4 | 4.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 1.4 | 14.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 1.3 | 5.4 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 1.2 | 6.9 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 1.2 | 4.6 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 1.1 | 5.3 | GO:2000173 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 1.0 | 3.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.7 | 2.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.6 | 2.5 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.6 | 4.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.6 | 7.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.5 | 7.5 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.5 | 2.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 5.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.5 | 4.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 4.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 | 5.6 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.4 | 1.2 | GO:0019541 | propionate metabolic process(GO:0019541) negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.4 | 1.5 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 5.0 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.3 | 4.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 3.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 1.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 5.6 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.3 | 1.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) negative regulation of telomere capping(GO:1904354) |
| 0.3 | 1.0 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 11.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 10.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.3 | 2.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 3.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.3 | 8.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 13.2 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.3 | 10.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.3 | 3.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 11.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.2 | 17.8 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.2 | 5.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 3.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 2.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 9.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 1.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 1.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.2 | 2.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 2.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 5.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 3.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 8.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 3.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 7.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 5.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 14.7 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 8.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 2.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 5.5 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 14.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 11.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 9.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 6.7 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 8.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 2.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 4.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 6.4 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 7.2 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 8.7 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 4.9 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 6.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 2.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.1 | GO:0071914 | prominosome(GO:0071914) |
| 2.2 | 17.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.2 | 4.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.9 | 2.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.9 | 8.6 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.8 | 14.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.8 | 10.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.7 | 8.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.7 | 13.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 8.2 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 5.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.6 | 5.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 3.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.5 | 3.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 4.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 2.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.4 | 5.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 5.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 5.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 26.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 2.2 | GO:0033263 | CORVET complex(GO:0033263) clathrin complex(GO:0071439) |
| 0.2 | 6.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 8.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 1.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 3.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 8.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 11.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 2.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 3.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 12.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 30.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 3.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 15.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 6.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 34.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 12.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 6.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 5.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 5.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 10.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 9.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 3.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 8.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.6 | 7.8 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 2.1 | 12.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 1.6 | 14.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.4 | 4.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.4 | 5.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 1.3 | 10.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.3 | 8.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.1 | 11.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.8 | 4.8 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.7 | 2.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.6 | 7.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.6 | 1.9 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 7.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.6 | 4.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 0.6 | 5.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.6 | 7.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 5.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 13.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.4 | 3.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.3 | 1.0 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.3 | 3.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 10.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.3 | 5.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 5.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 6.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 5.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 5.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 4.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 8.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 5.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 7.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 10.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 3.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 5.9 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 24.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 11.8 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 3.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 9.4 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 3.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 16.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 10.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 50.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 6.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 24.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 20.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 5.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 5.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 10.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 4.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 24.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.5 | 17.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 3.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 8.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 3.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 7.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 6.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 7.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 8.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 7.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 5.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 5.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 7.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 10.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.8 | 11.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.7 | 17.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.7 | 7.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.7 | 8.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.6 | 24.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.6 | 21.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 7.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 5.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 7.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 12.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 7.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 8.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 5.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 5.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 5.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 7.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 8.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 8.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 6.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |