Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
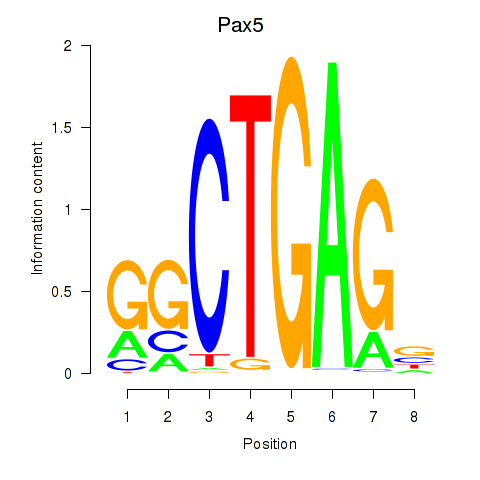
Results for Pax5
Z-value: 1.36
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSRNOG00000024729 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | rn6_v1_chr5_-_60191941_60191941 | 0.33 | 9.9e-10 | Click! |
Activity profile of Pax5 motif
Sorted Z-values of Pax5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_119441487 | 54.85 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr10_-_56167426 | 41.77 |
ENSRNOT00000013955
|
Efnb3
|
ephrin B3 |
| chr10_+_65586993 | 32.73 |
ENSRNOT00000086036
|
Aldoc
|
aldolase, fructose-bisphosphate C |
| chr10_-_15928169 | 32.21 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr13_-_90676629 | 28.98 |
ENSRNOT00000058143
|
Atp1a2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr1_-_93949187 | 28.66 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr3_-_72602548 | 28.36 |
ENSRNOT00000031745
|
Lrrc55
|
leucine rich repeat containing 55 |
| chr5_+_147323240 | 28.31 |
ENSRNOT00000047152
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr20_-_4823475 | 26.95 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr8_-_98738446 | 26.25 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr16_-_18974729 | 24.32 |
ENSRNOT00000086424
ENSRNOT00000064060 |
Tmem38a
|
transmembrane protein 38a |
| chr1_-_265560386 | 24.13 |
ENSRNOT00000048592
|
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr1_+_262905570 | 23.95 |
ENSRNOT00000090765
|
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr11_-_83905889 | 22.87 |
ENSRNOT00000075606
|
Fam131a
|
family with sequence similarity 131, member A |
| chr11_-_82884660 | 22.84 |
ENSRNOT00000073526
|
LOC100912498
|
protein FAM131A-like |
| chr2_+_210977938 | 22.43 |
ENSRNOT00000074725
|
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr15_-_33775109 | 22.13 |
ENSRNOT00000033722
|
Jph4
|
junctophilin 4 |
| chr8_+_39305128 | 21.43 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr15_+_33600102 | 21.06 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr14_-_55081551 | 20.86 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr8_+_29453643 | 20.76 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr2_+_220298245 | 20.72 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chrX_+_70563570 | 20.43 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr10_-_90999506 | 20.38 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr5_-_144779212 | 20.37 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr4_-_169999873 | 19.93 |
ENSRNOT00000011697
|
Grin2b
|
glutamate ionotropic receptor NMDA type subunit 2B |
| chr11_+_80358211 | 19.91 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr9_-_21338582 | 18.77 |
ENSRNOT00000016940
|
Ptchd4
|
patched domain containing 4 |
| chr10_+_65586504 | 18.76 |
ENSRNOT00000015535
ENSRNOT00000046388 |
Aldoc
|
aldolase, fructose-bisphosphate C |
| chr15_+_33600337 | 18.65 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr9_-_85243001 | 18.63 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr2_+_188844073 | 18.60 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr6_-_137084739 | 18.42 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chrX_+_105239840 | 18.11 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chr10_-_74679858 | 18.02 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr13_-_49313940 | 17.93 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chrX_-_157028434 | 17.93 |
ENSRNOT00000080088
|
Plxnb3
|
plexin B3 |
| chr4_+_110700403 | 17.88 |
ENSRNOT00000092379
|
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chrX_-_15707436 | 17.54 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr17_-_10818835 | 17.51 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr1_-_5165859 | 17.04 |
ENSRNOT00000044325
ENSRNOT00000019319 |
Grm1
|
glutamate metabotropic receptor 1 |
| chr20_+_7279820 | 16.50 |
ENSRNOT00000090820
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr2_+_41157823 | 16.43 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr4_-_157381105 | 16.42 |
ENSRNOT00000021670
|
Gpr162
|
G protein-coupled receptor 162 |
| chr14_+_2194933 | 16.42 |
ENSRNOT00000061720
|
Cplx1
|
complexin 1 |
| chr6_+_41917132 | 16.16 |
ENSRNOT00000071387
|
Ntsr2
|
neurotensin receptor 2 |
| chr14_+_37116492 | 16.14 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr4_-_52350624 | 15.96 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr8_+_118333706 | 15.70 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr3_+_47995959 | 15.40 |
ENSRNOT00000082775
|
Slc4a10
|
solute carrier family 4 member 10 |
| chrX_+_23081125 | 15.27 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chr7_+_141346398 | 15.18 |
ENSRNOT00000077175
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr7_-_121029754 | 14.99 |
ENSRNOT00000004703
|
Nptxr
|
neuronal pentraxin receptor |
| chr4_+_35279063 | 14.91 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr4_-_157078130 | 14.86 |
ENSRNOT00000015570
|
Clstn3
|
calsyntenin 3 |
| chr16_-_64806050 | 14.53 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr14_+_115275894 | 14.38 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr11_+_75905443 | 14.33 |
ENSRNOT00000002650
|
Fgf12
|
fibroblast growth factor 12 |
| chr6_-_142585188 | 14.10 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr13_-_111581018 | 14.09 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr7_+_145117951 | 14.03 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr12_-_5773036 | 13.99 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr18_-_73873280 | 13.82 |
ENSRNOT00000075580
|
Rnf165
|
ring finger protein 165 |
| chr1_+_199449973 | 13.61 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr3_+_11629556 | 13.60 |
ENSRNOT00000074401
|
St6galnac6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr10_+_89089646 | 13.52 |
ENSRNOT00000027505
|
Cntnap1
|
contactin associated protein 1 |
| chr6_+_110624856 | 13.42 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr14_+_25589762 | 13.41 |
ENSRNOT00000043938
ENSRNOT00000067439 ENSRNOT00000002793 |
Epha5
|
EPH receptor A5 |
| chr13_+_89622632 | 13.09 |
ENSRNOT00000004713
|
Adamts4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
| chr7_+_70364813 | 12.93 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr15_-_44860604 | 12.73 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr4_+_64088900 | 12.62 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr15_-_95514259 | 12.62 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr19_+_55669626 | 12.22 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr13_-_49487892 | 12.10 |
ENSRNOT00000044972
|
Nfasc
|
neurofascin |
| chr8_+_27777179 | 11.88 |
ENSRNOT00000009825
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 |
| chr12_+_39554171 | 11.77 |
ENSRNOT00000024347
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr1_+_97632473 | 11.62 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr6_-_141488290 | 11.56 |
ENSRNOT00000067336
|
AABR07065792.1
|
|
| chr11_+_66713888 | 11.40 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr13_-_51992693 | 11.37 |
ENSRNOT00000008282
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr7_-_98709344 | 11.29 |
ENSRNOT00000064122
|
Tmem65
|
transmembrane protein 65 |
| chr3_-_160301552 | 11.28 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr14_+_8182383 | 10.87 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr1_-_170404056 | 10.84 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr8_-_115167486 | 10.79 |
ENSRNOT00000033018
|
Gpr62
|
G protein-coupled receptor 62 |
| chr13_+_92569785 | 10.77 |
ENSRNOT00000082700
|
Fmn2
|
formin 2 |
| chr6_-_141291347 | 10.76 |
ENSRNOT00000008333
|
AABR07065789.1
|
|
| chr10_+_83636518 | 10.73 |
ENSRNOT00000007352
|
Phospho1
|
phosphoethanolamine/phosphocholine phosphatase 1 |
| chr12_+_28212333 | 10.73 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr7_+_60099120 | 10.70 |
ENSRNOT00000007338
|
LOC100911101
|
leucine-rich repeat-containing protein 10-like |
| chr17_-_23923792 | 10.56 |
ENSRNOT00000018739
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr12_-_23954431 | 10.38 |
ENSRNOT00000001958
|
Mdh2
|
malate dehydrogenase 2 |
| chr4_+_180291389 | 10.29 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr12_+_2534212 | 10.24 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr4_-_103369224 | 10.14 |
ENSRNOT00000075709
|
AABR07061057.1
|
|
| chr8_+_65611570 | 10.12 |
ENSRNOT00000017147
|
Larp6
|
La ribonucleoprotein domain family, member 6 |
| chr5_+_76092287 | 10.06 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr5_+_148661070 | 9.99 |
ENSRNOT00000056229
|
Nkain1
|
Sodium/potassium transporting ATPase interacting 1 |
| chr9_-_55673704 | 9.95 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr10_+_45258887 | 9.95 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr7_+_2752680 | 9.93 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr20_+_5008508 | 9.87 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr7_-_119996824 | 9.76 |
ENSRNOT00000011079
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chrX_-_1714061 | 9.63 |
ENSRNOT00000011592
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr3_+_19495628 | 9.58 |
ENSRNOT00000077990
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr5_-_168734296 | 9.57 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr3_+_18970574 | 9.57 |
ENSRNOT00000088394
|
AABR07051665.1
|
|
| chrX_+_105239620 | 9.50 |
ENSRNOT00000085693
|
Drp2
|
dystrophin related protein 2 |
| chr8_-_117237229 | 9.46 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr16_-_81945127 | 9.43 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr13_-_51183269 | 9.37 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr12_-_30728655 | 9.21 |
ENSRNOT00000035881
|
Mmp17
|
matrix metallopeptidase 17 |
| chr18_+_30574627 | 9.20 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr1_+_221801524 | 9.20 |
ENSRNOT00000031227
|
Nrxn2
|
neurexin 2 |
| chr6_-_142676432 | 9.17 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr1_+_77994203 | 9.13 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr11_+_85430400 | 9.05 |
ENSRNOT00000083198
|
AABR07034729.1
|
|
| chr4_-_103646906 | 9.02 |
ENSRNOT00000047620
|
AABR07061078.1
|
|
| chr5_-_65073012 | 8.96 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chrX_+_105500173 | 8.94 |
ENSRNOT00000040476
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr11_+_85532526 | 8.83 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr15_+_4850122 | 8.82 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr20_-_2191640 | 8.82 |
ENSRNOT00000001016
|
Trim10
|
tripartite motif-containing 10 |
| chr5_+_159967839 | 8.81 |
ENSRNOT00000051317
|
Hspb7
|
heat shock protein family B (small) member 7 |
| chr13_+_71107465 | 8.80 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr11_-_27971359 | 8.79 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr6_-_142635763 | 8.77 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr1_-_57815038 | 8.74 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chr10_-_108691367 | 8.74 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr20_+_3558827 | 8.72 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_+_85508300 | 8.57 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr10_+_16635989 | 8.52 |
ENSRNOT00000028155
|
Nkx2-5
|
NK2 homeobox 5 |
| chr6_-_99870024 | 8.49 |
ENSRNOT00000010043
|
Rab15
|
RAB15, member RAS oncogene family |
| chr7_+_60087429 | 8.48 |
ENSRNOT00000073117
|
Lrrc10
|
leucine-rich repeat-containing 10 |
| chr18_+_4325875 | 8.43 |
ENSRNOT00000073771
|
Impact
|
impact RWD domain protein |
| chr1_-_261051498 | 8.31 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr10_-_15155412 | 8.26 |
ENSRNOT00000026536
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr2_+_184230459 | 8.25 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr6_-_143065639 | 8.12 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chr4_+_132137793 | 8.12 |
ENSRNOT00000014455
|
Gpr27
|
G protein-coupled receptor 27 |
| chr2_+_34186091 | 8.11 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr10_-_64657089 | 8.02 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr1_-_59347472 | 8.01 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr17_-_31813716 | 7.95 |
ENSRNOT00000074147
|
AABR07027451.1
|
|
| chr11_+_85618714 | 7.92 |
ENSRNOT00000074614
|
AC109901.1
|
|
| chr15_-_33752665 | 7.90 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr19_-_37427989 | 7.89 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr8_+_118066988 | 7.87 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr1_-_215846911 | 7.86 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr17_+_54181419 | 7.71 |
ENSRNOT00000023861
|
Kif5b
|
kinesin family member 5B |
| chr15_-_108898703 | 7.64 |
ENSRNOT00000067577
|
Zic5
|
Zic family member 5 |
| chr7_-_116255167 | 7.60 |
ENSRNOT00000038109
ENSRNOT00000041774 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr1_-_43638161 | 7.53 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chrX_-_2657155 | 7.52 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr2_+_187737770 | 7.48 |
ENSRNOT00000036143
ENSRNOT00000092611 ENSRNOT00000092620 |
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr10_+_90348299 | 7.48 |
ENSRNOT00000089616
|
Rundc3a
|
RUN domain containing 3A |
| chr11_+_85992696 | 7.42 |
ENSRNOT00000084433
|
AABR07034736.1
|
|
| chr1_+_79989019 | 7.42 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr5_+_127404450 | 7.40 |
ENSRNOT00000017575
|
Lrp8
|
LDL receptor related protein 8 |
| chr6_+_137997335 | 7.39 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr17_+_31441630 | 7.32 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr1_-_140860882 | 7.32 |
ENSRNOT00000067889
ENSRNOT00000024253 ENSRNOT00000079100 |
Mfge8
|
milk fat globule-EGF factor 8 protein |
| chr5_-_85123829 | 7.30 |
ENSRNOT00000007578
|
Brinp1
|
BMP/retinoic acid inducible neural specific 1 |
| chr15_+_32386816 | 7.08 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr6_-_140407307 | 7.07 |
ENSRNOT00000079087
|
AABR07065768.3
|
|
| chr6_-_142880382 | 7.07 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr8_+_45797315 | 7.06 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chrX_-_63961527 | 7.04 |
ENSRNOT00000071762
|
Gspt2
|
G1 to S phase transition 2 |
| chr2_+_187988925 | 7.00 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr3_+_19174027 | 6.99 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr1_-_14412807 | 6.94 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr2_+_184243976 | 6.88 |
ENSRNOT00000042146
|
Dear
|
dual endothelin 1, angiotensin II receptor |
| chr6_+_139523337 | 6.86 |
ENSRNOT00000090711
|
AABR07065699.4
|
|
| chr6_-_143537030 | 6.82 |
ENSRNOT00000071876
|
LOC100360610
|
rCG64220-like |
| chr16_+_23668595 | 6.81 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_103761881 | 6.81 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr15_+_52241801 | 6.75 |
ENSRNOT00000082639
|
Hr
|
HR, lysine demethylase and nuclear receptor corepressor |
| chr20_-_29738506 | 6.72 |
ENSRNOT00000000697
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr2_+_182796728 | 6.70 |
ENSRNOT00000032351
|
LOC100909434
|
protocadherin-16-like |
| chr11_-_1983513 | 6.64 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr5_+_142875773 | 6.57 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr12_-_21760292 | 6.56 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr10_-_94557764 | 6.52 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chrX_+_135470915 | 6.51 |
ENSRNOT00000009637
|
Slc25a14
|
solute carrier family 25 member 14 |
| chr13_-_51784639 | 6.45 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_139523495 | 6.41 |
ENSRNOT00000075467
|
AABR07065699.4
|
|
| chr20_+_22060224 | 6.37 |
ENSRNOT00000057992
|
Zfp365
|
zinc finger protein 365 |
| chr2_+_207930796 | 6.37 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr10_+_70262361 | 6.35 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_-_211831551 | 6.34 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr1_-_72335855 | 6.30 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr5_+_164808323 | 6.27 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr1_+_165237847 | 6.23 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr4_+_102403451 | 6.20 |
ENSRNOT00000071605
|
AABR07060995.1
|
|
| chr3_-_79018487 | 6.13 |
ENSRNOT00000080981
|
Olr736
|
olfactory receptor 736 |
| chr14_+_83724933 | 6.13 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chrX_+_14498119 | 6.12 |
ENSRNOT00000051135
|
Xk
|
X-linked Kx blood group |
| chr1_-_252550394 | 6.09 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_-_11494459 | 6.09 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 41.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 7.2 | 29.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 5.8 | 17.5 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 5.5 | 16.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 5.4 | 16.3 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 4.8 | 14.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 4.7 | 23.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 4.2 | 12.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 4.2 | 41.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 4.0 | 19.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 3.7 | 51.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.6 | 10.8 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 3.6 | 17.9 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 3.6 | 10.7 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 3.5 | 14.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.4 | 20.4 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 3.4 | 13.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 3.2 | 25.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 3.2 | 28.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 3.1 | 28.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 2.9 | 20.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 2.8 | 8.5 | GO:0003285 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) His-Purkinje system cell differentiation(GO:0060932) |
| 2.8 | 17.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 2.8 | 8.4 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 2.8 | 11.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 2.7 | 13.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 2.7 | 8.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 2.7 | 8.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.7 | 8.0 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 2.6 | 7.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.6 | 7.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.6 | 13.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 2.5 | 10.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 2.5 | 12.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.5 | 12.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.5 | 14.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.4 | 16.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 2.3 | 6.9 | GO:0034146 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 2.3 | 9.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 2.1 | 6.4 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 2.1 | 25.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.1 | 6.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 1.9 | 5.8 | GO:0061744 | motor behavior(GO:0061744) |
| 1.9 | 3.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.9 | 5.7 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 1.8 | 7.3 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 1.8 | 9.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 1.8 | 16.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.8 | 7.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.8 | 17.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 1.7 | 7.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.7 | 9.9 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.7 | 8.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.7 | 14.9 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.6 | 17.9 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 1.6 | 7.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.5 | 6.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 1.5 | 6.0 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.5 | 6.0 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining(GO:0097681) |
| 1.5 | 7.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.4 | 23.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 1.4 | 6.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 1.4 | 39.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.3 | 10.7 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 1.3 | 4.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.3 | 2.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.3 | 7.9 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 1.3 | 26.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.3 | 5.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.3 | 11.4 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 1.3 | 3.8 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.3 | 5.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.2 | 18.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.2 | 13.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.2 | 3.7 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 1.2 | 18.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.2 | 14.0 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 1.2 | 20.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 1.1 | 3.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.1 | 3.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.1 | 9.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.0 | 7.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.0 | 15.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.0 | 10.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.0 | 3.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 1.0 | 4.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.9 | 6.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.9 | 3.6 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.9 | 8.8 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.9 | 4.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.9 | 10.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.9 | 6.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 2.5 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.8 | 3.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.8 | 22.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.8 | 6.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.8 | 6.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.8 | 4.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.8 | 4.0 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.8 | 8.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.8 | 9.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.8 | 8.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.8 | 2.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.8 | 2.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.8 | 6.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 3.8 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.8 | 17.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.7 | 16.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.7 | 3.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 5.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 | 5.7 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.7 | 9.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.7 | 2.6 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.7 | 9.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.7 | 6.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.6 | 3.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.6 | 1.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 4.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.6 | 3.7 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.6 | 4.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.6 | 3.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.6 | 1.8 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 13.8 | GO:0060384 | innervation(GO:0060384) |
| 0.6 | 20.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.6 | 26.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.6 | 4.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.6 | 5.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.6 | 2.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.6 | 7.3 | GO:0007614 | short-term memory(GO:0007614) maternal behavior(GO:0042711) |
| 0.6 | 3.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.6 | 2.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.5 | 2.7 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.5 | 2.7 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.5 | 3.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 5.9 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.5 | 8.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 3.3 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.5 | 11.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.5 | 6.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.4 | 1.8 | GO:1990859 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.4 | 4.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 1.3 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 6.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 3.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.4 | 8.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.4 | 12.9 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.4 | 9.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 1.6 | GO:1903999 | defecation(GO:0030421) negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.4 | 1.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) multivesicular body assembly(GO:0036258) |
| 0.4 | 9.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 2.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 25.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.4 | 52.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 3.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 5.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 2.9 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 2.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 1.8 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.3 | 1.4 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.3 | 2.0 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.3 | 2.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 14.8 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.3 | 8.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 21.2 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.3 | 1.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 3.5 | GO:0032341 | aldosterone metabolic process(GO:0032341) aldosterone biosynthetic process(GO:0032342) |
| 0.3 | 6.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.3 | 5.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 4.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 6.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 2.1 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.3 | 0.9 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 3.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 3.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.3 | 39.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.3 | 3.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 18.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.3 | 2.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 2.7 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 0.8 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 3.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 3.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 5.8 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.2 | 4.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 1.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 2.9 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 24.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.2 | 5.6 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 10.6 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 3.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 0.4 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.2 | 0.7 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 6.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 23.3 | GO:0050808 | synapse organization(GO:0050808) |
| 0.2 | 3.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 1.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.2 | 4.0 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.2 | 5.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 2.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 4.8 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 0.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 0.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 4.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 2.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 0.7 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.2 | 1.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 1.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 1.8 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 1.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 6.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 4.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.3 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 4.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 1.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 7.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 3.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 2.7 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 1.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 4.8 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 3.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 8.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 4.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 4.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 2.8 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 1.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 3.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.4 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 20.2 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.1 | 0.6 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.8 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.4 | GO:0051701 | interaction with host(GO:0051701) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.2 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.2 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 17.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 5.1 | 20.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 4.6 | 9.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 4.3 | 17.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 3.9 | 11.8 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 3.3 | 16.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 2.7 | 10.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 2.7 | 16.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.6 | 15.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 2.5 | 22.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.4 | 12.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.4 | 29.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 2.1 | 10.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 2.1 | 12.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.9 | 44.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.8 | 7.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.7 | 28.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.5 | 37.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.5 | 8.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.3 | 4.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.3 | 18.6 | GO:0031045 | dense core granule(GO:0031045) |
| 1.2 | 6.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.2 | 12.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.1 | 17.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.0 | 9.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.0 | 11.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 1.0 | 3.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.9 | 1.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.9 | 90.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.9 | 12.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.9 | 12.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.9 | 7.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.8 | 4.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.8 | 10.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 5.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.6 | 3.5 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.6 | 4.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.6 | 17.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 7.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 25.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 46.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 34.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 10.4 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.4 | 7.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 69.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 3.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 2.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 15.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 9.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 6.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 4.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 6.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 3.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 28.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 19.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 1.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 5.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 4.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 9.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 1.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 29.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.2 | 3.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 15.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 3.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 16.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 4.4 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.2 | 7.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.2 | 23.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 7.6 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 15.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 7.3 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 4.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 3.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 21.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 3.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 72.9 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 5.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 0.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 4.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 3.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 30.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 7.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 3.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 5.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 15.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 3.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 24.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 10.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 4.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 8.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 51.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 6.1 | 30.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 5.1 | 15.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 5.0 | 15.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 5.0 | 19.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 4.8 | 14.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 4.2 | 12.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 4.0 | 16.2 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 4.0 | 11.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 3.5 | 14.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 3.4 | 34.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 3.1 | 22.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 3.0 | 9.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 3.0 | 27.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 2.9 | 23.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 2.7 | 13.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 2.7 | 15.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 2.6 | 10.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 2.5 | 7.6 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 2.4 | 14.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 2.1 | 19.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 2.1 | 14.9 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 2.0 | 9.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.9 | 7.7 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.9 | 17.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.9 | 20.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.8 | 16.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 1.7 | 17.0 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 1.7 | 8.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.6 | 55.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.5 | 9.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.5 | 6.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.5 | 8.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.5 | 5.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.3 | 13.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.3 | 7.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.3 | 6.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.3 | 9.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.2 | 8.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.2 | 6.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.1 | 4.6 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 1.1 | 3.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.1 | 24.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 1.1 | 8.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.1 | 10.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 3.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.0 | 5.0 | GO:0043559 | insulin binding(GO:0043559) |
| 1.0 | 9.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.9 | 3.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.9 | 5.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.9 | 16.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.9 | 2.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.9 | 5.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.9 | 20.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.9 | 3.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.8 | 43.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.8 | 12.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.8 | 30.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.8 | 3.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 4.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.7 | 7.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 13.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.7 | 4.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.7 | 10.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.7 | 18.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.7 | 8.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 3.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 6.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.6 | 21.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.6 | 4.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.6 | 7.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.6 | 11.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.6 | 13.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 5.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 6.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.5 | 11.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 1.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 3.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 6.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.5 | 8.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 12.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 4.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 2.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 4.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.4 | 5.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 4.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 8.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 53.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.4 | 10.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 14.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 4.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 7.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 7.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 8.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 2.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.3 | 2.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 6.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 7.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 28.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 1.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 10.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.3 | 2.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 5.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 3.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 1.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 1.3 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.3 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 5.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.8 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 6.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 8.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
| 0.2 | 15.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 4.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 3.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 8.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 2.9 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 21.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 14.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 1.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 2.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 18.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.2 | 1.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 2.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 8.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 3.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 18.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 106.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 4.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 8.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 1.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 11.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 4.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 14.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 5.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 8.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.7 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 5.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.0 | GO:0005267 | potassium channel activity(GO:0005267) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 30.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.9 | 21.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.8 | 18.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.8 | 44.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.7 | 8.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 4.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.6 | 7.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 13.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.5 | 7.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 16.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 11.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.4 | 11.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.4 | 13.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 13.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.4 | 8.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 4.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 3.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 8.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 13.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 6.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 2.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.2 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 4.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 7.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 4.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 9.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 12.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.7 | 33.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 1.3 | 51.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 20.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.1 | 19.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.0 | 18.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.9 | 20.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.8 | 11.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.8 | 15.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.8 | 7.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.8 | 17.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 10.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 8.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.7 | 29.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.7 | 14.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.7 | 9.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.6 | 24.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 14.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.6 | 7.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 14.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 12.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 6.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 7.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 7.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 7.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 6.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 9.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.4 | 5.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 13.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 8.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 31.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 13.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 3.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.3 | 5.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 8.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 6.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 20.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 4.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 3.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.2 | 1.8 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 2.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 5.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 7.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 5.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 6.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 16.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 6.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 6.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |