Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pax4
Z-value: 0.54
Transcription factors associated with Pax4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax4
|
ENSRNOG00000008020 | paired box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax4 | rn6_v1_chr4_-_55740627_55740627 | -0.21 | 1.4e-04 | Click! |
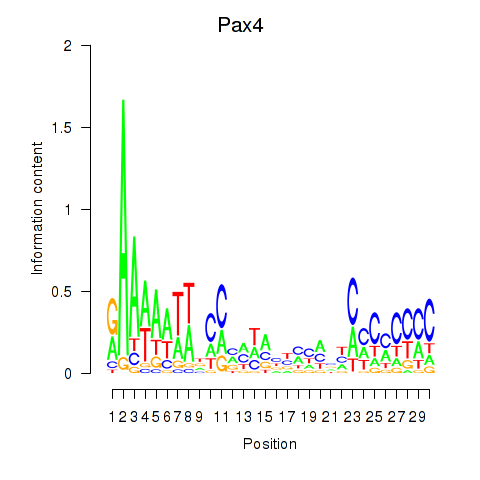
Activity profile of Pax4 motif
Sorted Z-values of Pax4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_3904 | 22.25 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_81639872 | 18.97 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chrM_+_9870 | 16.83 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chrM_+_10160 | 15.70 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr19_+_37476095 | 14.08 |
ENSRNOT00000092794
ENSRNOT00000023130 |
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr3_-_51297852 | 13.28 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr1_-_248376750 | 12.71 |
ENSRNOT00000089073
|
Gldc
|
glycine decarboxylase |
| chr5_+_6373583 | 12.18 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chrM_+_11736 | 10.05 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr1_+_105285419 | 8.60 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr4_-_176720012 | 8.57 |
ENSRNOT00000017965
|
Ldhb
|
lactate dehydrogenase B |
| chr2_+_252452269 | 8.02 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr3_-_170955399 | 7.73 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr15_+_43298794 | 7.47 |
ENSRNOT00000012736
|
Adra1a
|
adrenoceptor alpha 1A |
| chr8_+_87630916 | 6.58 |
ENSRNOT00000016428
|
Myo6
|
myosin VI |
| chr4_-_158704170 | 6.55 |
ENSRNOT00000082009
|
Ntf3
|
neurotrophin 3 |
| chr14_-_103321270 | 6.48 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr3_+_114176309 | 6.44 |
ENSRNOT00000023350
|
Sord
|
sorbitol dehydrogenase |
| chr7_-_119689938 | 6.36 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr14_+_99919485 | 6.24 |
ENSRNOT00000006087
|
Egfr
|
epidermal growth factor receptor |
| chr1_-_188407132 | 5.90 |
ENSRNOT00000073233
|
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr6_-_103313074 | 5.55 |
ENSRNOT00000083677
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr13_-_80775230 | 5.41 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr12_-_44999074 | 5.25 |
ENSRNOT00000079177
ENSRNOT00000038293 |
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr8_-_54990604 | 5.20 |
ENSRNOT00000059192
|
Tex12
|
testis expressed 12 |
| chr7_-_101140308 | 5.19 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr1_-_164308317 | 5.09 |
ENSRNOT00000022983
|
Serpinh1
|
serpin family H member 1 |
| chr10_-_56558487 | 4.97 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr7_+_97559841 | 4.43 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chrX_+_15035569 | 4.26 |
ENSRNOT00000006491
ENSRNOT00000078392 |
Porcn
|
porcupine homolog (Drosophila) |
| chr18_-_71829881 | 4.11 |
ENSRNOT00000088662
|
Ctif
|
cap binding complex dependent translation initiation factor |
| chr18_-_57245666 | 4.04 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr16_-_62373253 | 3.98 |
ENSRNOT00000034325
|
Tex15
|
testis expressed 15, meiosis and synapsis associated |
| chr20_+_5049496 | 3.82 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_37018459 | 3.65 |
ENSRNOT00000087297
|
Eea1
|
early endosome antigen 1 |
| chr8_-_53901358 | 3.40 |
ENSRNOT00000047666
ENSRNOT00000042281 |
Ncam1
|
neural cell adhesion molecule 1 |
| chrX_+_105537602 | 3.31 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr5_-_172307431 | 3.26 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr3_-_113438801 | 3.18 |
ENSRNOT00000091665
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chrX_-_102510007 | 3.17 |
ENSRNOT00000082854
|
AABR07040494.1
|
|
| chr20_-_3818045 | 2.75 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr11_-_57993548 | 2.66 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr3_-_162579201 | 2.64 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chrX_+_16337102 | 2.55 |
ENSRNOT00000003954
|
Ccnb3
|
cyclin B3 |
| chr10_-_25845634 | 2.49 |
ENSRNOT00000004269
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr1_+_80092403 | 2.30 |
ENSRNOT00000078336
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr12_-_37411525 | 2.26 |
ENSRNOT00000087698
|
Tctn2
|
tectonic family member 2 |
| chr1_+_252894663 | 2.16 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr4_-_99810270 | 2.06 |
ENSRNOT00000078123
|
Ptcd3
|
Pentatricopeptide repeat domain 3 |
| chr14_-_43143973 | 2.00 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr17_+_45801528 | 1.76 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr17_-_27969433 | 1.76 |
ENSRNOT00000073967
|
Nrn1
|
neuritin 1 |
| chrX_-_29825439 | 1.73 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr8_+_12823155 | 1.67 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chr15_+_27346731 | 1.66 |
ENSRNOT00000046317
|
Olr1619
|
olfactory receptor 1619 |
| chr8_-_67040005 | 1.62 |
ENSRNOT00000038641
|
Glce
|
glucuronic acid epimerase |
| chr15_-_6587367 | 1.55 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr7_+_122203532 | 1.38 |
ENSRNOT00000025382
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr7_-_114573900 | 1.35 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chr18_+_30527705 | 1.33 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr9_+_20765296 | 1.29 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr3_+_160047296 | 1.28 |
ENSRNOT00000051590
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr9_-_10047507 | 1.25 |
ENSRNOT00000072118
|
Alkbh7
|
alkB homolog 7 |
| chr1_-_221370322 | 1.21 |
ENSRNOT00000028431
|
Capn1
|
calpain 1 |
| chrX_+_128416722 | 1.18 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr20_-_4401610 | 1.13 |
ENSRNOT00000091468
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr9_-_10471176 | 1.10 |
ENSRNOT00000089164
|
Safb
|
scaffold attachment factor B |
| chr2_+_188561429 | 1.07 |
ENSRNOT00000076458
ENSRNOT00000027867 |
Krtcap2
|
keratinocyte associated protein 2 |
| chr3_-_103043741 | 1.06 |
ENSRNOT00000052294
|
Olr775
|
olfactory receptor 775 |
| chr4_-_99746560 | 1.04 |
ENSRNOT00000012021
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr7_+_9279620 | 0.99 |
ENSRNOT00000011225
|
Olr1064
|
olfactory receptor 1064 |
| chr6_-_114488880 | 0.99 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr3_-_77800137 | 0.95 |
ENSRNOT00000071319
|
AABR07052831.1
|
|
| chr4_+_87353254 | 0.89 |
ENSRNOT00000073670
|
Vom1r67
|
vomeronasal 1 receptor 67 |
| chr8_+_19972793 | 0.86 |
ENSRNOT00000068066
|
Olr1159
|
olfactory receptor 1159 |
| chr8_-_18062971 | 0.81 |
ENSRNOT00000045739
|
Olr1139
|
olfactory receptor 1139 |
| chr3_-_148932878 | 0.81 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr18_+_25613831 | 0.81 |
ENSRNOT00000091040
|
Tslp
|
thymic stromal lymphopoietin |
| chr10_-_43912900 | 0.79 |
ENSRNOT00000029524
|
Olr1414
|
olfactory receptor 1414 |
| chrX_-_70428364 | 0.76 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr7_-_6754349 | 0.72 |
ENSRNOT00000085061
|
Olr956
|
olfactory receptor 956 |
| chr7_+_117605050 | 0.71 |
ENSRNOT00000047380
|
Slc52a2
|
solute carrier family 52 member 2 |
| chr8_-_70436028 | 0.70 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr13_-_53108713 | 0.63 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr4_+_71795768 | 0.57 |
ENSRNOT00000031921
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr20_-_1878126 | 0.50 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr6_+_28235695 | 0.33 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr10_-_87351030 | 0.32 |
ENSRNOT00000034889
ENSRNOT00000091152 |
Krt20
|
keratin 20 |
| chr2_-_250862419 | 0.27 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr1_+_273854248 | 0.26 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr8_-_17524839 | 0.14 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr1_-_169845487 | 0.11 |
ENSRNOT00000039989
|
Olr179
|
olfactory receptor 179 |
| chr7_-_138512952 | 0.08 |
ENSRNOT00000009187
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr3_+_102859815 | 0.06 |
ENSRNOT00000037906
|
Olr771
|
olfactory receptor 771 |
| chrX_-_123486814 | 0.01 |
ENSRNOT00000016993
|
RGD1564541
|
similar to hypothetical protein FLJ22965 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 19.0 | GO:0097037 | heme export(GO:0097037) |
| 3.2 | 12.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.6 | 7.7 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 2.5 | 7.5 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 2.1 | 6.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.7 | 8.6 | GO:0036233 | glycine import(GO:0036233) |
| 1.6 | 37.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.6 | 6.6 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 1.4 | 5.6 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) mesendoderm development(GO:0048382) regulation of intracellular mRNA localization(GO:1904580) |
| 1.4 | 5.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 1.1 | 3.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.9 | 6.2 | GO:1900019 | astrocyte activation(GO:0048143) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.9 | 13.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.9 | 14.1 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.8 | 8.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.7 | 5.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.7 | 7.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.7 | 2.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 2.6 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.6 | 5.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.6 | 26.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.5 | 2.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 1.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.4 | 1.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.4 | 2.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.4 | 8.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 9.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 | 1.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 2.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 6.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 2.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 3.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 1.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 5.0 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 3.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 4.0 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.2 | 2.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 1.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 3.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 3.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 4.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 4.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 4.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 4.3 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 2.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 19.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 4.2 | 12.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 1.6 | 6.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.3 | 64.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.8 | 2.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 3.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.4 | 8.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 5.2 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 5.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 3.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 6.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 2.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 4.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 7.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 17.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 5.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 4.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 6.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 4.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 21.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.9 | 8.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 2.5 | 7.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.4 | 64.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.1 | 8.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.6 | 6.2 | GO:0048408 | epidermal growth factor-activated receptor activity(GO:0005006) epidermal growth factor binding(GO:0048408) |
| 1.3 | 3.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.1 | 8.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.1 | 3.2 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 1.0 | 5.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.9 | 2.7 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.8 | 6.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.8 | 18.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.6 | 7.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 12.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.6 | 5.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 2.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 5.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 13.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 5.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 0.8 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 3.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 4.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 6.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 5.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 6.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.3 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 6.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 4.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 6.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 6.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 5.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 16.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 4.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 8.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 8.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 6.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 5.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 5.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 5.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 7.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 19.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 9.2 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |