Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
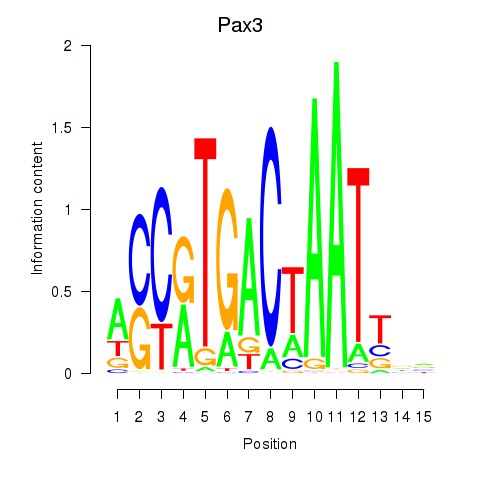
Results for Pax3
Z-value: 0.39
Transcription factors associated with Pax3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax3
|
ENSRNOG00000013670 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax3 | rn6_v1_chr9_-_84101172_84101172 | -0.16 | 4.6e-03 | Click! |
Activity profile of Pax3 motif
Sorted Z-values of Pax3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_39958239 | 12.83 |
ENSRNOT00000050368
|
Myl2
|
myosin light chain 2 |
| chr2_+_201289357 | 12.65 |
ENSRNOT00000067358
|
Tbx15
|
T-box 15 |
| chr2_+_208750356 | 9.20 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr1_+_261281543 | 8.36 |
ENSRNOT00000018890
|
Ankrd2
|
ankyrin repeat domain 2 |
| chr15_+_28295368 | 8.09 |
ENSRNOT00000013786
|
Slc39a2
|
solute carrier family 39 member 2 |
| chr9_-_79530821 | 6.77 |
ENSRNOT00000085619
|
Mreg
|
melanoregulin |
| chr5_+_119903507 | 6.04 |
ENSRNOT00000033933
|
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr15_+_57290849 | 5.90 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chrX_-_110230610 | 5.80 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr9_-_10170428 | 5.62 |
ENSRNOT00000073048
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr4_+_120156659 | 4.64 |
ENSRNOT00000050584
|
Dnajb8
|
DnaJ heat shock protein family (Hsp40) member B8 |
| chr4_-_84131030 | 4.63 |
ENSRNOT00000012196
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr12_-_18227991 | 4.48 |
ENSRNOT00000072986
|
AABR07035501.1
|
|
| chr2_-_90568486 | 3.88 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr2_-_173836854 | 3.75 |
ENSRNOT00000074278
|
Wdr49
|
WD repeat domain 49 |
| chr2_-_206222248 | 3.57 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr3_+_172195844 | 3.50 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr4_+_14109864 | 3.44 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr6_-_94932806 | 3.35 |
ENSRNOT00000006346
|
Ccdc175
|
coiled-coil domain containing 175 |
| chr2_-_132301073 | 3.09 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr10_+_56710965 | 2.90 |
ENSRNOT00000087121
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_+_76833761 | 2.74 |
ENSRNOT00000042650
|
Uts2b
|
urotensin 2B |
| chrY_-_1398030 | 2.43 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr1_-_227965386 | 2.41 |
ENSRNOT00000028491
|
Ms4a2
|
membrane spanning 4-domains A2 |
| chr17_+_15845931 | 2.33 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr6_-_42630983 | 2.23 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr15_-_6587367 | 2.22 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr8_+_116311078 | 2.11 |
ENSRNOT00000037363
|
Rassf1
|
Ras association domain family member 1 |
| chr5_+_162246217 | 1.52 |
ENSRNOT00000046115
|
Oog1
|
oogenesin 1 |
| chr5_+_126698582 | 1.35 |
ENSRNOT00000012427
|
Cdcp2
|
CUB domain containing protein 2 |
| chr1_+_172264473 | 1.27 |
ENSRNOT00000050651
|
Olr246
|
olfactory receptor 246 |
| chr20_-_11620945 | 1.17 |
ENSRNOT00000079725
|
Krtap12-2
|
keratin associated protein 12-2 |
| chr18_+_28409955 | 1.17 |
ENSRNOT00000026989
|
Paip2
|
poly(A) binding protein interacting protein 2 |
| chr17_-_44853430 | 1.15 |
ENSRNOT00000085156
|
Olr1654
|
olfactory receptor 1654 |
| chr11_-_78456200 | 0.98 |
ENSRNOT00000067251
ENSRNOT00000036179 |
Tp63
|
tumor protein p63 |
| chr11_-_84037938 | 0.98 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr4_+_148544958 | 0.98 |
ENSRNOT00000050694
|
Olr825
|
olfactory receptor 825 |
| chr1_+_225690756 | 0.94 |
ENSRNOT00000027498
|
Psbpc2
|
prostatic steroid-binding protein C2 |
| chr3_-_78942535 | 0.83 |
ENSRNOT00000008924
|
Olr731
|
olfactory receptor 731 |
| chr8_-_36842370 | 0.77 |
ENSRNOT00000074011
|
Pate3
|
prostate and testis expressed 3 |
| chr12_+_16899025 | 0.75 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr12_-_21761487 | 0.74 |
ENSRNOT00000082910
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr1_+_127604197 | 0.73 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
| chr15_-_28092023 | 0.72 |
ENSRNOT00000090835
|
Eddm3b
|
epididymal protein 3B |
| chr1_-_80221710 | 0.70 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr3_-_173166369 | 0.67 |
ENSRNOT00000090602
|
AABR07054887.1
|
|
| chr3_-_73484676 | 0.64 |
ENSRNOT00000040041
|
Olr481
|
olfactory receptor 481 |
| chr7_-_116963281 | 0.64 |
ENSRNOT00000082058
ENSRNOT00000012306 |
Tsta3
|
tissue specific transplantation antigen P35B |
| chr3_-_167270512 | 0.62 |
ENSRNOT00000075542
|
AABR07054708.1
|
|
| chr13_+_92465894 | 0.61 |
ENSRNOT00000086988
|
LOC103693685
|
olfactory receptor 6P1 |
| chr16_+_50152008 | 0.60 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr8_-_21341431 | 0.58 |
ENSRNOT00000044249
|
Olr1192
|
olfactory receptor 1192 |
| chr18_+_25962855 | 0.50 |
ENSRNOT00000078822
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_+_5167685 | 0.40 |
ENSRNOT00000046109
|
Lcn4
|
lipocalin 4 |
| chr5_-_22799349 | 0.37 |
ENSRNOT00000076204
|
Asph
|
aspartate-beta-hydroxylase |
| chr3_+_20993205 | 0.36 |
ENSRNOT00000010465
|
Olr419
|
olfactory receptor 419 |
| chr10_-_86645529 | 0.30 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr16_-_20511818 | 0.30 |
ENSRNOT00000026567
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_172728264 | 0.27 |
ENSRNOT00000075297
|
Olr267
|
olfactory receptor 267 |
| chr15_+_28050975 | 0.23 |
ENSRNOT00000050944
|
LOC103693384
|
epididymal secretory protein E3-beta-like |
| chr5_-_29870735 | 0.21 |
ENSRNOT00000012963
|
Ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chrX_+_86126157 | 0.19 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr10_-_35800120 | 0.17 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr6_-_105316024 | 0.17 |
ENSRNOT00000009483
|
Med6
|
mediator complex subunit 6 |
| chr17_-_45746753 | 0.17 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr8_+_21305250 | 0.17 |
ENSRNOT00000050206
|
Olr1193
|
olfactory receptor 1193 |
| chr6_-_138662365 | 0.16 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr4_-_168656673 | 0.15 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr5_-_137515690 | 0.14 |
ENSRNOT00000049593
|
LOC100912421
|
olfactory receptor 2D2-like |
| chr10_-_60942351 | 0.06 |
ENSRNOT00000026926
|
Olr1509
|
olfactory receptor 1509 |
| chr1_+_79122608 | 0.03 |
ENSRNOT00000023663
|
Ceacam3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 2.0 | 5.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.8 | 9.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 8.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) regulation of myoblast proliferation(GO:2000291) |
| 0.5 | 5.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 4.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 6.8 | GO:0032401 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.3 | 8.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 1.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 12.7 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.2 | 0.6 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.2 | 2.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 2.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0070671 | positive regulation of T-helper 1 cell differentiation(GO:0045627) response to interleukin-12(GO:0070671) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 2.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 6.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 2.7 | GO:0008217 | regulation of blood pressure(GO:0008217) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.4 | 12.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 2.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 8.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 6.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 16.7 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.5 | 9.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.2 | 4.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.7 | 5.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 12.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 2.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 12.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 8.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 5.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 4.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.5 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 12.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |