Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
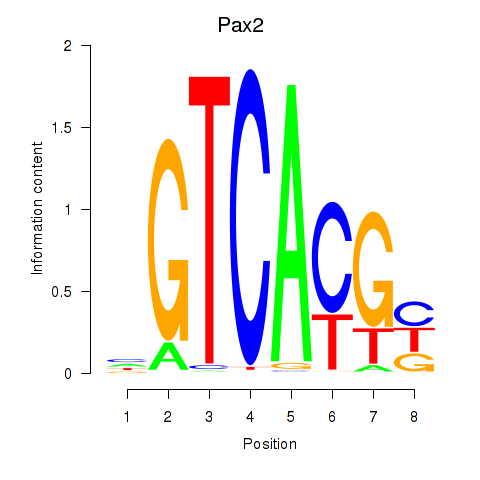
Results for Pax2
Z-value: 0.36
Transcription factors associated with Pax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax2
|
ENSRNOG00000014253 | paired box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax2 | rn6_v1_chr1_+_264504591_264504591 | 0.25 | 4.9e-06 | Click! |
Activity profile of Pax2 motif
Sorted Z-values of Pax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_161298962 | 9.45 |
ENSRNOT00000066028
|
Ctsa
|
cathepsin A |
| chr4_+_33638709 | 7.72 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chrX_-_54303729 | 6.82 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr7_+_142912316 | 6.57 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_+_55940533 | 5.69 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr19_+_24329023 | 5.56 |
ENSRNOT00000065558
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr1_+_263367266 | 5.39 |
ENSRNOT00000054714
|
Nkx2-3
|
NK2 homeobox 3 |
| chr7_-_12741296 | 4.92 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr5_-_164844586 | 4.61 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chr7_+_53878610 | 4.18 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr10_-_66602987 | 4.00 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr11_+_60613882 | 3.85 |
ENSRNOT00000002853
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr18_+_27657628 | 3.84 |
ENSRNOT00000026303
|
Egr1
|
early growth response 1 |
| chr6_-_108976489 | 3.73 |
ENSRNOT00000007350
|
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr20_+_6288267 | 3.47 |
ENSRNOT00000000627
|
Srsf3
|
serine and arginine rich splicing factor 3 |
| chr10_-_56276764 | 3.35 |
ENSRNOT00000049048
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr1_+_154377247 | 3.27 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr3_-_161322289 | 3.07 |
ENSRNOT00000022594
|
Pltp
|
phospholipid transfer protein |
| chr13_-_42263024 | 3.01 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr10_-_104637823 | 3.01 |
ENSRNOT00000010826
|
Wbp2
|
WW domain binding protein 2 |
| chr2_+_188516582 | 2.98 |
ENSRNOT00000074727
|
Gba
|
glucosylceramidase beta |
| chr19_-_37525762 | 2.94 |
ENSRNOT00000023606
|
Atp6v0d1
|
ATPase H+ transporting V0 subunit D1 |
| chr15_-_27855999 | 2.91 |
ENSRNOT00000013225
|
Tmem55b
|
transmembrane protein 55B |
| chr5_+_147375350 | 2.48 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr5_-_147375009 | 2.38 |
ENSRNOT00000009436
|
S100pbp
|
S100P binding protein |
| chr5_+_78222504 | 2.30 |
ENSRNOT00000019544
|
Slc31a1
|
solute carrier family 31 member 1 |
| chr1_+_154377447 | 2.23 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_-_70498992 | 2.13 |
ENSRNOT00000067774
ENSRNOT00000079327 |
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr10_+_10725819 | 2.11 |
ENSRNOT00000004159
|
Glyr1
|
glyoxylate reductase 1 homolog |
| chr11_-_60613718 | 2.01 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr3_+_163570532 | 1.94 |
ENSRNOT00000010054
|
Arfgef2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr10_-_74070266 | 1.65 |
ENSRNOT00000005987
|
Cltc
|
clathrin heavy chain |
| chr3_-_164239250 | 1.58 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chr2_-_219693629 | 1.48 |
ENSRNOT00000020907
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr9_+_16647598 | 1.33 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr10_-_62287189 | 1.26 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr2_+_2456735 | 1.24 |
ENSRNOT00000032974
|
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr10_-_10725655 | 1.18 |
ENSRNOT00000061236
|
Ubn1
|
ubinuclein 1 |
| chr1_-_102849430 | 1.13 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr8_-_103298927 | 1.09 |
ENSRNOT00000046873
ENSRNOT00000011609 |
U2surp
|
U2 snRNP-associated SURP domain containing |
| chr2_+_240396152 | 1.04 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr15_-_20822740 | 0.99 |
ENSRNOT00000012957
|
Bmp4
|
bone morphogenetic protein 4 |
| chr12_+_37593874 | 0.98 |
ENSRNOT00000057902
|
Sbno1
|
strawberry notch homolog 1 |
| chr9_+_16647922 | 0.98 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr7_-_92882068 | 0.97 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr3_-_161299024 | 0.91 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr10_+_20591432 | 0.83 |
ENSRNOT00000059780
|
Pank3
|
pantothenate kinase 3 |
| chr12_+_37594185 | 0.80 |
ENSRNOT00000088787
|
Sbno1
|
strawberry notch homolog 1 |
| chr6_+_28663602 | 0.72 |
ENSRNOT00000005402
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr6_+_108167716 | 0.72 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr10_+_14492844 | 0.69 |
ENSRNOT00000023615
|
Clcn7
|
chloride voltage-gated channel 7 |
| chr13_+_26172243 | 0.65 |
ENSRNOT00000003840
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr12_+_19303387 | 0.63 |
ENSRNOT00000001820
|
Cops6
|
COP9 signalosome subunit 6 |
| chr8_-_48674748 | 0.46 |
ENSRNOT00000014127
|
Hmbs
|
hydroxymethylbilane synthase |
| chr5_-_62001196 | 0.46 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr1_-_274107138 | 0.39 |
ENSRNOT00000078670
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr11_-_38457373 | 0.34 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr1_+_103172987 | 0.34 |
ENSRNOT00000018688
|
Tmem86a
|
transmembrane protein 86A |
| chr2_+_207108552 | 0.30 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr6_-_109205004 | 0.19 |
ENSRNOT00000010512
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr1_-_219450451 | 0.08 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr16_-_9658484 | 0.05 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr10_-_56850085 | 0.01 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.8 | 5.5 | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 1.6 | 9.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.5 | 7.7 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 1.3 | 3.8 | GO:2000182 | interleukin-1 biosynthetic process(GO:0042222) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 1.1 | 6.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.1 | 5.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.0 | 4.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.0 | 3.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.0 | 3.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.8 | 3.0 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.6 | 2.3 | GO:1902861 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.5 | 1.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 1.0 | GO:1902462 | intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) cardiac jelly development(GO:1905072) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.3 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 2.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 1.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 2.0 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 3.0 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 5.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 2.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 2.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 3.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 3.5 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 3.9 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 4.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 1.9 | GO:0032280 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.2 | 2.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 2.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 2.9 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 1.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 6.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 5.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 5.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 8.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 2.3 | 6.8 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.0 | 3.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.7 | 2.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 5.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 2.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.5 | 2.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 2.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 3.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 2.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 5.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 1.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 6.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 3.9 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 3.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 2.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 7.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 4.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 3.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.5 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 3.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 9.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 7.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 1.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 6.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 2.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 7.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |