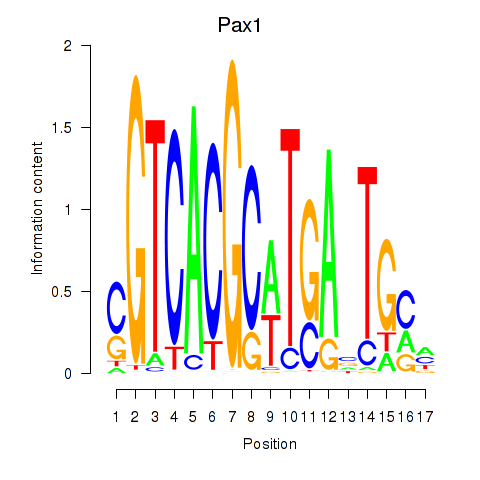
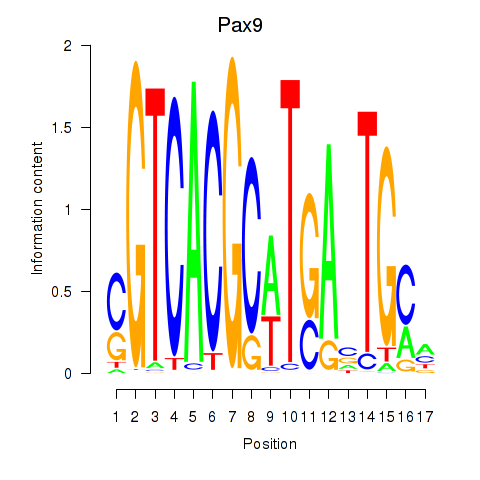
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pax1_Pax9
Z-value: 0.54
Transcription factors associated with Pax1_Pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax1
|
ENSRNOG00000024882 | paired box 1 |
|
Pax9
|
ENSRNOG00000008826 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax1 | rn6_v1_chr3_+_141577504_141577504 | -0.34 | 4.2e-10 | Click! |
| Pax9 | rn6_v1_chr6_+_77608624_77608624 | -0.24 | 9.6e-06 | Click! |
Activity profile of Pax1_Pax9 motif
Sorted Z-values of Pax1_Pax9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_179307095 | 30.64 |
ENSRNOT00000021193
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr2_+_220298245 | 20.21 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chrX_+_118084890 | 19.19 |
ENSRNOT00000065293
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr4_+_79557854 | 13.59 |
ENSRNOT00000013145
|
Npy
|
neuropeptide Y |
| chr20_+_13778178 | 13.12 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr4_+_79573998 | 13.08 |
ENSRNOT00000074351
|
LOC100912228
|
pro-neuropeptide Y-like |
| chr18_-_1946840 | 11.77 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr3_+_128828331 | 10.71 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr15_+_67092304 | 10.57 |
ENSRNOT00000040244
|
AABR07018646.1
|
|
| chr1_-_18511695 | 10.29 |
ENSRNOT00000075402
|
AABR07000583.1
|
|
| chr8_+_70603249 | 9.80 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr1_-_220502272 | 8.96 |
ENSRNOT00000041238
|
Klc2
|
kinesin light chain 2 |
| chr1_-_178196294 | 8.88 |
ENSRNOT00000067170
|
Btbd10
|
BTB domain containing 10 |
| chr5_-_57896475 | 7.85 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr12_+_7208850 | 6.94 |
ENSRNOT00000001219
|
Katnal1
|
katanin catalytic subunit A1 like 1 |
| chr5_-_39689824 | 6.82 |
ENSRNOT00000046713
|
AABR07047606.1
|
|
| chr20_+_7484550 | 6.74 |
ENSRNOT00000044395
|
Anks1a
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr1_+_18353812 | 6.73 |
ENSRNOT00000029394
|
AABR07000581.1
|
|
| chr1_-_19522171 | 6.67 |
ENSRNOT00000045386
|
AABR07000595.1
|
|
| chr4_+_174692331 | 6.52 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr3_-_15433252 | 5.48 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr9_-_65737538 | 4.81 |
ENSRNOT00000014939
|
Trak2
|
trafficking kinesin protein 2 |
| chr20_+_7484800 | 4.02 |
ENSRNOT00000081604
|
Anks1a
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr16_-_24788740 | 3.96 |
ENSRNOT00000018952
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr6_+_26390689 | 3.77 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr11_+_43329700 | 3.47 |
ENSRNOT00000060892
|
Olr1540
|
olfactory receptor 1540 |
| chr1_+_198682230 | 3.15 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr8_+_126411829 | 3.00 |
ENSRNOT00000013450
|
Azi2
|
5-azacytidine induced 2 |
| chrX_-_157172068 | 2.85 |
ENSRNOT00000087962
|
Dusp9
|
dual specificity phosphatase 9 |
| chr2_+_124150224 | 2.58 |
ENSRNOT00000078650
|
Spata5
|
spermatogenesis associated 5 |
| chr5_-_50068706 | 2.50 |
ENSRNOT00000084643
|
Orc3
|
origin recognition complex, subunit 3 |
| chr1_-_165997751 | 2.38 |
ENSRNOT00000050227
|
P2ry6
|
pyrimidinergic receptor P2Y6 |
| chrX_-_83788325 | 2.23 |
ENSRNOT00000037149
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr16_-_24788456 | 1.88 |
ENSRNOT00000079412
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr12_+_37709860 | 1.30 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr17_-_79676499 | 1.05 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr3_+_2490518 | 1.04 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr8_+_43355060 | 0.93 |
ENSRNOT00000043206
|
Olr1309
|
olfactory receptor 1309 |
| chr9_-_53732858 | 0.88 |
ENSRNOT00000029204
|
Nemp2
|
nuclear envelope integral membrane protein 2 |
| chr6_+_53401109 | 0.79 |
ENSRNOT00000014763
|
Twist1
|
twist family bHLH transcription factor 1 |
| chr9_-_17880706 | 0.67 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr18_+_64114933 | 0.63 |
ENSRNOT00000022364
|
Mc5r
|
melanocortin 5 receptor |
| chr6_+_135270871 | 0.48 |
ENSRNOT00000051685
ENSRNOT00000079048 |
Zfp839
|
zinc finger protein 839 |
| chr3_+_147609095 | 0.32 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr7_+_2435553 | 0.19 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr3_+_2877293 | 0.15 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr9_-_100306194 | 0.14 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr8_-_55276070 | 0.04 |
ENSRNOT00000041555
|
LOC108351703
|
60S ribosomal protein L27a-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax1_Pax9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 30.6 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 6.4 | 19.2 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 5.3 | 26.7 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 2.7 | 10.8 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 1.6 | 4.8 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 1.0 | 3.0 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.8 | 2.5 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 5.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 6.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 20.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.4 | 5.8 | GO:0030432 | peristalsis(GO:0030432) |
| 0.4 | 2.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 10.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.4 | 1.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.3 | 2.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 8.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 3.8 | GO:0061525 | hindgut development(GO:0061525) |
| 0.3 | 11.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.3 | 0.8 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 9.0 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 2.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 7.8 | GO:0043623 | cellular protein complex assembly(GO:0043623) |
| 0.0 | 6.5 | GO:0061458 | reproductive system development(GO:0061458) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 7.8 | GO:0002177 | manchette(GO:0002177) |
| 0.6 | 3.8 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.6 | 9.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 10.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 4.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 39.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 13.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 8.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 6.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 5.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 30.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 6.4 | 19.2 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 2.0 | 11.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.8 | 20.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.2 | 5.8 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.9 | 26.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.8 | 2.4 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.7 | 2.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.5 | 10.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.5 | 6.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 6.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 4.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 13.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 7.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 10.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 9.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 30.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 10.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 30.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.4 | 19.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 9.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 10.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.2 | 2.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 2.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 20.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |