Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
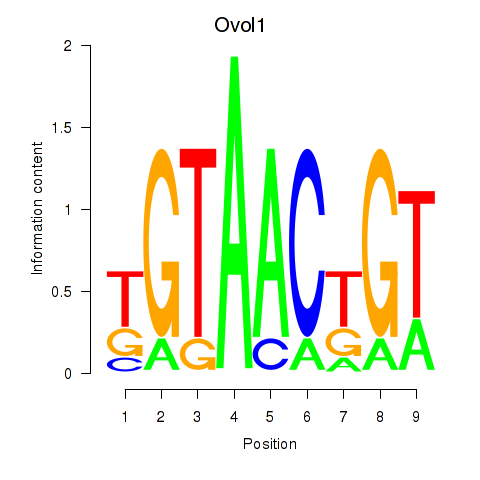
Results for Ovol1
Z-value: 0.24
Transcription factors associated with Ovol1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ovol1
|
ENSRNOG00000020669 | ovo like transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ovol1 | rn6_v1_chr1_-_220938814_220938814 | -0.18 | 9.7e-04 | Click! |
Activity profile of Ovol1 motif
Sorted Z-values of Ovol1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_44327551 | 8.63 |
ENSRNOT00000083939
|
Gramd1b
|
GRAM domain containing 1B |
| chr1_+_17602281 | 4.78 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr6_-_60124274 | 4.10 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr4_+_138441332 | 2.94 |
ENSRNOT00000090847
|
Cntn4
|
contactin 4 |
| chr3_+_95715193 | 2.43 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr14_-_17582823 | 2.35 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr2_-_250778269 | 2.27 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr14_+_23480462 | 1.78 |
ENSRNOT00000002755
|
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr2_-_250805445 | 1.44 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr7_-_140617721 | 1.12 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr15_-_28155291 | 0.97 |
ENSRNOT00000050820
|
Rnase2
|
ribonuclease A family member 2 |
| chr13_-_74331214 | 0.83 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr3_+_102859815 | 0.70 |
ENSRNOT00000037906
|
Olr771
|
olfactory receptor 771 |
| chr3_+_77921705 | 0.55 |
ENSRNOT00000064006
|
AC132028.1
|
|
| chr3_+_75768699 | 0.35 |
ENSRNOT00000071912
|
AABR07052798.1
|
|
| chr3_+_102553317 | 0.22 |
ENSRNOT00000043765
|
Olr753
|
olfactory receptor 753 |
| chr3_+_77584990 | 0.21 |
ENSRNOT00000046926
|
Olr664
|
olfactory receptor 664 |
| chr1_+_79981355 | 0.04 |
ENSRNOT00000020134
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr18_+_30036887 | 0.03 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ovol1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.2 | 1.8 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.9 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.2 | 2.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 3.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.4 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |