Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
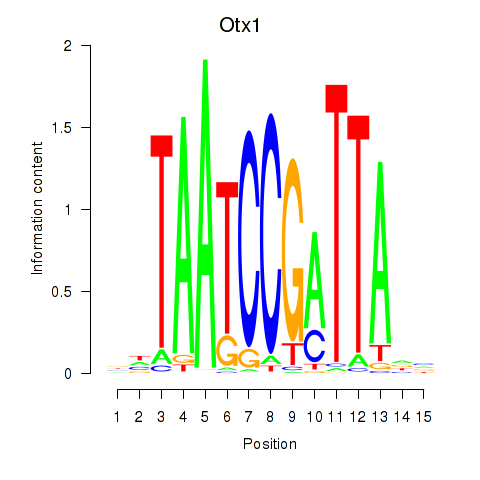
Results for Otx1
Z-value: 0.74
Transcription factors associated with Otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Otx1
|
ENSRNOG00000059469 | orthodenticle homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx1 | rn6_v1_chr14_-_106864892_106864892 | 0.35 | 6.7e-11 | Click! |
Activity profile of Otx1 motif
Sorted Z-values of Otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_106753592 | 38.92 |
ENSRNOT00000006930
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr11_+_57505005 | 22.30 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr2_-_265300868 | 21.95 |
ENSRNOT00000066024
ENSRNOT00000016073 ENSRNOT00000033502 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr1_+_27476375 | 18.42 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr11_+_60072727 | 17.64 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr3_+_56862691 | 17.46 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chrX_-_157286936 | 16.52 |
ENSRNOT00000078100
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr9_+_12114977 | 16.38 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr6_-_2311781 | 14.44 |
ENSRNOT00000084171
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr11_-_4397361 | 14.22 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr2_+_66940057 | 13.78 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chrX_-_56765893 | 13.28 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr13_-_39643361 | 12.49 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr15_+_19547871 | 12.22 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr6_-_127319362 | 11.06 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr2_+_102685513 | 10.51 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr2_+_198797159 | 10.45 |
ENSRNOT00000056225
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr12_-_10391270 | 9.85 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr9_+_116652530 | 9.29 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr9_-_114327767 | 9.28 |
ENSRNOT00000085481
|
AABR07068674.1
|
|
| chr11_+_57207656 | 8.62 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr18_-_67224566 | 8.62 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr10_-_66848388 | 8.44 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr16_-_39476384 | 8.43 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr1_+_40529045 | 8.39 |
ENSRNOT00000026564
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr18_+_30496318 | 8.05 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr3_-_162059524 | 7.70 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr9_+_62291809 | 7.63 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr1_-_214414763 | 7.52 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr19_+_14835822 | 7.32 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr1_+_65522118 | 7.29 |
ENSRNOT00000058563
|
Mzf1
|
myeloid zinc finger 1 |
| chr18_+_27047382 | 7.17 |
ENSRNOT00000027691
ENSRNOT00000090264 |
Apc
|
APC, WNT signaling pathway regulator |
| chr4_+_9981958 | 7.03 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr1_-_755645 | 6.76 |
ENSRNOT00000073546
|
Vom2r6
|
vomeronasal 2 receptor, 6 |
| chr14_-_46153212 | 6.56 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr8_+_64481172 | 6.40 |
ENSRNOT00000015332
|
Pkm
|
pyruvate kinase, muscle |
| chr12_-_8418966 | 6.35 |
ENSRNOT00000085869
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr6_-_104409005 | 6.34 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr10_+_45258887 | 6.28 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr2_-_156807305 | 6.26 |
ENSRNOT00000080578
|
AABR07011031.1
|
uncharacterized protein LOC302022 |
| chr9_+_49837868 | 5.61 |
ENSRNOT00000021999
|
Gpr45
|
G protein-coupled receptor 45 |
| chr8_+_129205931 | 5.61 |
ENSRNOT00000082377
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr14_+_3882398 | 5.53 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr6_-_114476723 | 5.37 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr6_-_67085390 | 5.31 |
ENSRNOT00000083277
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chrX_-_17252877 | 5.12 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chr11_+_45339366 | 4.91 |
ENSRNOT00000002241
|
Tmem30c
|
transmembrane protein 30C |
| chr8_+_82038967 | 4.64 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr6_+_92864123 | 4.48 |
ENSRNOT00000087540
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_+_70776046 | 4.38 |
ENSRNOT00000040403
|
Prss1
|
protease, serine 1 |
| chr4_+_87026530 | 4.35 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr12_-_54885 | 4.18 |
ENSRNOT00000090447
|
AABR07034833.1
|
|
| chr10_+_56381813 | 4.14 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chrX_+_984798 | 4.11 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr8_+_126975833 | 4.09 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr7_+_12006710 | 3.87 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr16_+_39144972 | 3.83 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr16_+_39353283 | 3.78 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr4_+_117962319 | 3.78 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr5_+_139385429 | 3.63 |
ENSRNOT00000078622
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr8_+_58347736 | 3.55 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_-_217620420 | 3.49 |
ENSRNOT00000092783
|
Cttn
|
cortactin |
| chr4_+_88119838 | 3.42 |
ENSRNOT00000073173
|
Vom1r82
|
vomeronasal 1 receptor 82 |
| chr18_-_55771730 | 3.33 |
ENSRNOT00000026426
|
Smim3
|
small integral membrane protein 3 |
| chr8_-_116855062 | 3.26 |
ENSRNOT00000050344
|
Rnf123
|
ring finger protein 123 |
| chr5_+_156396695 | 3.25 |
ENSRNOT00000067631
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr3_-_93693690 | 3.24 |
ENSRNOT00000086745
|
Nat10
|
N-acetyltransferase 10 |
| chr9_-_65442257 | 3.24 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr16_+_39145230 | 3.20 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr5_+_70528688 | 3.19 |
ENSRNOT00000036799
|
Fktn
|
fukutin |
| chr3_+_17009089 | 3.11 |
ENSRNOT00000048829
|
LOC100361052
|
rCG64257-like |
| chr12_+_31536951 | 3.09 |
ENSRNOT00000086078
|
Rimbp2
|
RIMS binding protein 2 |
| chrM_-_14061 | 3.09 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_72464492 | 3.08 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr19_+_359271 | 3.03 |
ENSRNOT00000086313
|
AABR07042623.1
|
|
| chr2_+_260444227 | 3.01 |
ENSRNOT00000064249
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr6_+_100337226 | 3.00 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr20_-_14573519 | 2.90 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr11_-_43099412 | 2.80 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chrM_+_14136 | 2.77 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr19_-_9931930 | 2.77 |
ENSRNOT00000085144
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr8_-_104995725 | 2.75 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr4_+_88207124 | 2.72 |
ENSRNOT00000082459
|
Vom1r84
|
vomeronasal 1 receptor 84 |
| chr20_+_37587687 | 2.69 |
ENSRNOT00000001050
|
Msl3l2
|
male-specific lethal 3-like 2 (Drosophila) |
| chr1_-_70485888 | 2.49 |
ENSRNOT00000020514
|
Olr7
|
olfactory receptor 7 |
| chr3_-_21684132 | 2.47 |
ENSRNOT00000012364
|
Zbtb6
|
zinc finger and BTB domain containing 6 |
| chr14_+_36071376 | 2.46 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr2_+_93520734 | 2.41 |
ENSRNOT00000083909
|
Snx16
|
sorting nexin 16 |
| chr19_+_26066127 | 2.38 |
ENSRNOT00000064289
|
Rtbdn
|
retbindin |
| chr7_-_62162453 | 2.38 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chrM_+_10160 | 2.33 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr10_+_57218087 | 2.29 |
ENSRNOT00000089853
|
Mink1
|
misshapen-like kinase 1 |
| chr11_+_43329700 | 2.21 |
ENSRNOT00000060892
|
Olr1540
|
olfactory receptor 1540 |
| chr1_+_224998172 | 2.17 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chrM_+_11736 | 2.12 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_-_12945192 | 2.11 |
ENSRNOT00000082529
|
Cyth3
|
cytohesin 3 |
| chr3_-_76696107 | 2.04 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr8_-_117820413 | 1.97 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr1_-_102699442 | 1.93 |
ENSRNOT00000056109
|
Tph1
|
tryptophan hydroxylase 1 |
| chr14_-_71973419 | 1.89 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr4_+_144985880 | 1.84 |
ENSRNOT00000009464
|
Thumpd3
|
THUMP domain containing 3 |
| chr3_+_9822580 | 1.82 |
ENSRNOT00000010786
ENSRNOT00000093718 |
Usp20
|
ubiquitin specific peptidase 20 |
| chr17_-_14373983 | 1.82 |
ENSRNOT00000071112
|
AABR07027098.1
|
|
| chr8_-_57830504 | 1.74 |
ENSRNOT00000017683
|
Ddx10
|
DEAD-box helicase 10 |
| chr7_-_29171783 | 1.71 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr10_-_48038647 | 1.64 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr3_+_73115052 | 1.62 |
ENSRNOT00000090772
|
Olr454
|
olfactory receptor 454 |
| chr4_+_2711385 | 1.61 |
ENSRNOT00000035996
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr3_+_130008885 | 1.60 |
ENSRNOT00000070972
|
Slx4ip
|
SLX4 interacting protein |
| chr3_+_111826297 | 1.60 |
ENSRNOT00000081897
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr1_-_201942344 | 1.57 |
ENSRNOT00000027956
|
RGD1305014
|
similar to RIKEN cDNA 2310057M21 |
| chr6_-_143537030 | 1.56 |
ENSRNOT00000071876
|
LOC100360610
|
rCG64220-like |
| chr4_-_117589464 | 1.50 |
ENSRNOT00000021167
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr20_-_1099336 | 1.47 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr1_+_69853553 | 1.45 |
ENSRNOT00000042086
|
Zfp954
|
zinc finger protein 954 |
| chr1_-_90035522 | 1.43 |
ENSRNOT00000028672
|
Uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr3_+_121596791 | 1.39 |
ENSRNOT00000024694
|
Ttl
|
tubulin tyrosine ligase |
| chr1_+_230450529 | 1.39 |
ENSRNOT00000045383
|
Olr371
|
olfactory receptor 371 |
| chr7_+_123963381 | 1.39 |
ENSRNOT00000036586
|
Serhl2
|
serine hydrolase-like 2 |
| chr13_+_94888078 | 1.36 |
ENSRNOT00000077995
ENSRNOT00000005647 ENSRNOT00000077345 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr2_+_112914375 | 1.33 |
ENSRNOT00000092737
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr4_-_121822781 | 1.24 |
ENSRNOT00000072082
|
Vom1r95
|
vomeronasal 1 receptor 95 |
| chr17_+_43900450 | 1.23 |
ENSRNOT00000023622
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr8_-_18762922 | 1.23 |
ENSRNOT00000008423
|
Olr1122
|
olfactory receptor 1122 |
| chrX_+_152196129 | 1.19 |
ENSRNOT00000087679
|
Magea4
|
melanoma antigen, family A, 4 |
| chrX_+_123751089 | 1.18 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr4_+_38240728 | 1.16 |
ENSRNOT00000047029
|
Phf14
|
PHD finger protein 14 |
| chr13_+_67545430 | 1.16 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr5_+_6373583 | 1.16 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr5_+_159612762 | 1.16 |
ENSRNOT00000012147
|
Spata21
|
spermatogenesis associated 21 |
| chr7_-_17520780 | 1.15 |
ENSRNOT00000029672
|
RGD1560291
|
similar to NACHT, leucine rich repeat and PYD containing 4A |
| chr13_+_51218468 | 1.14 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chr7_-_70467915 | 1.14 |
ENSRNOT00000088995
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr7_-_129919946 | 1.12 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr1_-_228263198 | 1.09 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr7_-_12519154 | 1.09 |
ENSRNOT00000093376
ENSRNOT00000077681 |
Gpx4
|
glutathione peroxidase 4 |
| chr1_-_149683357 | 1.08 |
ENSRNOT00000077975
|
Olr17
|
olfactory receptor 17 |
| chrM_+_9870 | 1.01 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr4_+_87312766 | 0.99 |
ENSRNOT00000052126
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr4_-_38240848 | 0.98 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr14_+_45033309 | 0.98 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
| chr7_+_97760134 | 0.96 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr1_+_80954858 | 0.93 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr1_+_229416489 | 0.90 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr1_-_155955173 | 0.87 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr3_+_11587941 | 0.87 |
ENSRNOT00000071505
|
Dpm2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr1_-_149633029 | 0.85 |
ENSRNOT00000048861
|
LOC100910049
|
olfactory receptor 14A2-like |
| chr18_-_45380730 | 0.84 |
ENSRNOT00000037904
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr1_-_63457134 | 0.82 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr5_-_160619650 | 0.81 |
ENSRNOT00000089981
|
Tmem51
|
transmembrane protein 51 |
| chr3_+_143151739 | 0.79 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chr8_+_40285637 | 0.75 |
ENSRNOT00000015369
|
Olr1199
|
olfactory receptor 1199 |
| chr4_+_1640318 | 0.75 |
ENSRNOT00000083649
|
Olr1249
|
olfactory receptor 1249 |
| chr9_-_15306465 | 0.75 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr1_+_67742949 | 0.73 |
ENSRNOT00000049350
|
Vom1r41
|
vomeronasal 1 receptor 41 |
| chr1_+_72580424 | 0.68 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr1_-_72129269 | 0.68 |
ENSRNOT00000049179
|
Vom1r35
|
vomeronasal 1 receptor 35 |
| chr18_+_30909490 | 0.68 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr17_+_39544281 | 0.67 |
ENSRNOT00000022871
|
Prl2b1
|
Prolactin family 2, subfamily b, member 1 |
| chr3_+_67870111 | 0.61 |
ENSRNOT00000012628
|
Nup35
|
nucleoporin 35 |
| chr1_-_229927937 | 0.61 |
ENSRNOT00000080145
|
Olr349
|
olfactory receptor 349 |
| chr11_-_14160697 | 0.60 |
ENSRNOT00000081096
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr3_+_23301455 | 0.60 |
ENSRNOT00000019405
|
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr3_+_16610086 | 0.59 |
ENSRNOT00000046231
|
LOC100361009
|
rCG64257-like |
| chr17_-_63879166 | 0.59 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr12_-_24724997 | 0.59 |
ENSRNOT00000025560
|
Abhd11
|
abhydrolase domain containing 11 |
| chr10_-_76039964 | 0.58 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr17_-_54808483 | 0.56 |
ENSRNOT00000075683
|
AABR07028049.1
|
|
| chr16_+_8823872 | 0.56 |
ENSRNOT00000027186
|
Drgx
|
dorsal root ganglia homeobox |
| chr1_-_167606343 | 0.54 |
ENSRNOT00000025003
|
Olr40
|
olfactory receptor 40 |
| chr3_-_151724654 | 0.52 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr3_-_152259156 | 0.51 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chr12_-_41497610 | 0.50 |
ENSRNOT00000001860
|
Iqcd
|
IQ motif containing D |
| chr4_-_164900857 | 0.49 |
ENSRNOT00000078690
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr8_+_116336441 | 0.48 |
ENSRNOT00000090123
ENSRNOT00000021590 |
Nat6
Hyal3
|
N-acetyltransferase 6 hyaluronoglucosaminidase 3 |
| chr3_+_102859815 | 0.46 |
ENSRNOT00000037906
|
Olr771
|
olfactory receptor 771 |
| chr8_-_1450138 | 0.45 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr4_+_106323089 | 0.43 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr3_+_16571602 | 0.42 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr11_+_28945575 | 0.41 |
ENSRNOT00000061600
ENSRNOT00000090364 |
AABR07033579.3
|
|
| chr1_+_75298364 | 0.41 |
ENSRNOT00000018981
|
Vom1r61
|
vomeronasal 1 receptor 61 |
| chr3_-_76926151 | 0.39 |
ENSRNOT00000049517
|
Olr641
|
olfactory receptor 641 |
| chr7_-_116955148 | 0.38 |
ENSRNOT00000087328
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chr3_+_138174054 | 0.37 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr4_+_87827582 | 0.36 |
ENSRNOT00000077211
|
LOC108348088
|
putative vomeronasal receptor-like protein 4 |
| chrX_-_123731294 | 0.36 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr3_+_75834770 | 0.34 |
ENSRNOT00000043371
|
Olr582
|
olfactory receptor 582 |
| chr3_-_75879716 | 0.32 |
ENSRNOT00000007553
|
Olr584
|
olfactory receptor 584 |
| chr3_+_125320680 | 0.28 |
ENSRNOT00000028891
|
LOC681292
|
hypothetical protein LOC681292 |
| chr20_-_28783589 | 0.27 |
ENSRNOT00000071663
|
Sept10
|
septin 10 |
| chr3_-_16441030 | 0.27 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chrX_+_134389417 | 0.26 |
ENSRNOT00000075721
|
LOC103694516
|
high mobility group protein B4-like |
| chr9_-_71798265 | 0.24 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chrX_+_43745604 | 0.23 |
ENSRNOT00000040825
|
LOC100362173
|
rCG36365-like |
| chr7_-_18793289 | 0.23 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr8_-_43249887 | 0.22 |
ENSRNOT00000060094
|
Olr1305
|
olfactory receptor 1305 |
| chr8_+_43628182 | 0.21 |
ENSRNOT00000090359
|
Olr1320
|
olfactory receptor 1320 |
| chr1_-_169892850 | 0.20 |
ENSRNOT00000037868
|
Olr183
|
olfactory receptor 183 |
| chr2_+_196594303 | 0.20 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chrX_+_13992064 | 0.20 |
ENSRNOT00000036543
|
LOC100363125
|
rCG42854-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 3.5 | 17.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.8 | 8.4 | GO:0015942 | formate metabolic process(GO:0015942) |
| 2.2 | 8.6 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 2.1 | 16.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.9 | 38.9 | GO:0061548 | ganglion development(GO:0061548) |
| 1.8 | 7.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 1.8 | 7.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 1.5 | 10.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 1.2 | 4.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.9 | 9.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.8 | 3.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.8 | 2.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.7 | 3.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.6 | 3.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.6 | 6.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 5.4 | GO:0042447 | phenol-containing compound catabolic process(GO:0019336) hormone catabolic process(GO:0042447) |
| 0.6 | 7.6 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.6 | 5.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.5 | 9.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.5 | 7.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.5 | 2.8 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.4 | 3.5 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.4 | 18.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 1.2 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.3 | 8.4 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.3 | 1.0 | GO:0042496 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.3 | 1.9 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 4.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 1.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 14.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 3.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 2.8 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.2 | 8.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 4.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 1.6 | GO:0060710 | chorio-allantoic fusion(GO:0060710) chorion development(GO:0060717) |
| 0.2 | 7.2 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 1.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.7 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 18.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 2.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 2.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 15.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 1.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.6 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 1.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 4.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 19.7 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.1 | 1.4 | GO:0061387 | regulation of extent of cell growth(GO:0061387) |
| 0.0 | 3.1 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.5 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 5.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 2.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.6 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.4 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 2.3 | 60.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.5 | 4.6 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.1 | 8.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.0 | 3.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.8 | 2.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.6 | 3.5 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.5 | 1.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 17.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 14.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 1.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 1.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.3 | 0.9 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 2.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 8.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 3.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 12.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 4.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 6.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.7 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 3.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 18.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 22.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 5.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.4 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.7 | 8.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.7 | 8.4 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.6 | 6.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.5 | 7.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.2 | 12.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.2 | 7.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.1 | 13.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.8 | 16.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 7.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 7.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 14.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 5.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.3 | 38.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.3 | 1.0 | GO:0042498 | Toll-like receptor 2 binding(GO:0035663) diacyl lipopeptide binding(GO:0042498) |
| 0.3 | 8.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 0.9 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 18.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 4.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 3.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.4 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 3.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 4.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 4.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 3.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 30.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.8 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 1.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 2.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 8.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 21.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 8.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 6.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 6.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 3.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 5.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 6.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 1.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.7 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 7.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 38.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.0 | 28.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 14.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.8 | 16.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.7 | 17.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 8.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.5 | 7.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.5 | 7.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 4.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 8.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |