Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Osr2_Osr1
Z-value: 0.59
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Osr2
|
ENSRNOG00000011136 | odd-skipped related transciption factor 2 |
|
Osr1
|
ENSRNOG00000004210 | odd-skipped related transciption factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr1 | rn6_v1_chr6_+_35314444_35314444 | -0.19 | 6.2e-04 | Click! |
| Osr2 | rn6_v1_chr7_+_74047814_74047814 | -0.16 | 5.4e-03 | Click! |
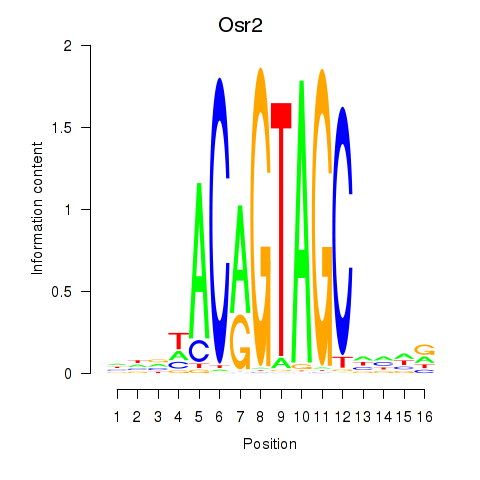
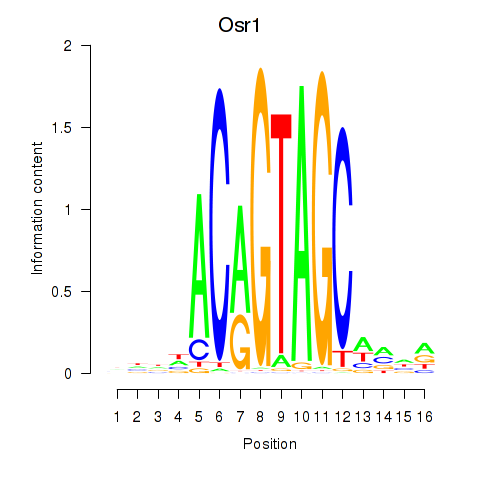
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_181486090 | 37.27 |
ENSRNOT00000075453
|
AABR07062535.1
|
|
| chr4_-_50200328 | 21.48 |
ENSRNOT00000060530
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr6_+_8284878 | 14.08 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr4_+_68849033 | 10.15 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr14_+_2050483 | 9.20 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr9_+_9721105 | 9.00 |
ENSRNOT00000073042
ENSRNOT00000075494 |
C3
|
complement C3 |
| chr1_+_154131926 | 8.28 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr17_-_9762813 | 8.19 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr2_+_235264219 | 7.97 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr2_-_5577369 | 7.48 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_+_3411380 | 6.92 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr2_-_192671059 | 6.88 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr7_-_145154131 | 6.57 |
ENSRNOT00000055271
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr17_+_76306585 | 6.51 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr8_+_117117430 | 6.09 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr15_+_18451144 | 6.06 |
ENSRNOT00000010260
|
Acox2
|
acyl-CoA oxidase 2 |
| chr3_+_119805941 | 5.92 |
ENSRNOT00000018584
|
Adra2b
|
adrenoceptor alpha 2B |
| chr13_+_89805962 | 5.81 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr1_+_48320802 | 5.71 |
ENSRNOT00000082903
|
AC114389.1
|
|
| chr20_-_31736212 | 5.55 |
ENSRNOT00000000662
|
RGD1305587
|
similar to RIKEN cDNA 2010107G23 |
| chr5_-_134526089 | 5.44 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr18_+_59748444 | 5.41 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_+_69634890 | 5.19 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr4_-_123557501 | 5.18 |
ENSRNOT00000075042
ENSRNOT00000085966 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr1_+_70222964 | 5.15 |
ENSRNOT00000050180
|
Peg3
|
paternally expressed 3 |
| chr1_+_248132090 | 5.08 |
ENSRNOT00000022056
|
Il33
|
interleukin 33 |
| chr5_-_135025084 | 5.04 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr3_-_151724654 | 5.02 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr13_+_89643881 | 4.92 |
ENSRNOT00000004735
|
B4galt3
|
beta-1,4-galactosyltransferase 3 |
| chr1_+_166564664 | 4.82 |
ENSRNOT00000090959
|
Pde2a
|
phosphodiesterase 2A |
| chr3_-_152259156 | 4.68 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chr10_-_59892960 | 4.65 |
ENSRNOT00000084432
|
Aspa
|
aspartoacylase |
| chr1_+_91363492 | 4.63 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr18_-_48384645 | 4.61 |
ENSRNOT00000023485
|
Ppic
|
peptidylprolyl isomerase C |
| chr8_+_48571323 | 4.55 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr18_+_32273770 | 4.53 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr2_-_35104963 | 4.38 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr6_-_42473738 | 4.32 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr6_+_10348308 | 4.28 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chrX_+_32232142 | 4.26 |
ENSRNOT00000047354
|
Ca5b
|
carbonic anhydrase 5B |
| chr18_+_16616937 | 4.24 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr3_-_150108898 | 4.24 |
ENSRNOT00000022914
|
Pxmp4
|
peroxisomal membrane protein 4 |
| chr15_-_18675431 | 4.18 |
ENSRNOT00000080794
|
Abhd6
|
abhydrolase domain containing 6 |
| chrX_-_75566481 | 4.12 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr5_+_152708775 | 4.10 |
ENSRNOT00000022713
|
Paqr7
|
progestin and adipoQ receptor family member 7 |
| chr10_+_68173369 | 4.01 |
ENSRNOT00000037072
|
Tmem98
|
transmembrane protein 98 |
| chr10_+_58342393 | 3.83 |
ENSRNOT00000010358
|
Wscd1
|
WSC domain containing 1 |
| chr19_-_40925660 | 3.74 |
ENSRNOT00000023645
|
Mtss1l
|
MTSS1L, I-BAR domain containing |
| chr2_+_58724855 | 3.68 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr8_-_117366096 | 3.68 |
ENSRNOT00000027411
|
Wdr6
|
WD repeat domain 6 |
| chr2_+_247299433 | 3.65 |
ENSRNOT00000050330
|
Unc5c
|
unc-5 netrin receptor C |
| chr1_-_173682226 | 3.57 |
ENSRNOT00000020137
|
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr7_-_102298522 | 3.56 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr14_+_71533460 | 3.56 |
ENSRNOT00000084274
|
Prom1
|
prominin 1 |
| chr11_-_82810014 | 3.56 |
ENSRNOT00000083539
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr1_+_266380973 | 3.54 |
ENSRNOT00000080509
|
Wbp1l
|
WW domain binding protein 1-like |
| chr1_+_162817611 | 3.53 |
ENSRNOT00000091952
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr4_-_171591882 | 3.51 |
ENSRNOT00000009328
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chrX_+_68782872 | 3.48 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr14_+_89253373 | 3.45 |
ENSRNOT00000035922
|
LOC688553
|
hypothetical protein LOC688553 |
| chr4_+_180123905 | 3.43 |
ENSRNOT00000083173
|
Rassf8
|
Ras association domain family member 8 |
| chr18_-_58423196 | 3.31 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chrX_+_35599258 | 3.30 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr11_+_69484293 | 3.30 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr1_+_31545631 | 3.27 |
ENSRNOT00000018336
|
Sdha
|
succinate dehydrogenase complex flavoprotein subunit A |
| chr18_+_31444472 | 3.23 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr1_-_87468288 | 3.19 |
ENSRNOT00000042207
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr1_+_256101903 | 3.18 |
ENSRNOT00000022384
|
Hhex
|
hematopoietically expressed homeobox |
| chr2_+_45104305 | 3.18 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr13_+_35554964 | 3.16 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr9_+_90857308 | 3.07 |
ENSRNOT00000073993
|
LOC100911572
|
collagen alpha-3(IV) chain-like |
| chr10_-_70788309 | 3.07 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr15_+_93634820 | 2.99 |
ENSRNOT00000093318
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr1_+_141832774 | 2.98 |
ENSRNOT00000073302
|
Fuom
|
fucose mutarotase |
| chrX_+_156355376 | 2.95 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr9_-_11080155 | 2.94 |
ENSRNOT00000072830
|
Fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr5_+_154310453 | 2.92 |
ENSRNOT00000013322
|
Gale
|
UDP-galactose-4-epimerase |
| chr11_-_31892531 | 2.91 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr11_+_71222600 | 2.79 |
ENSRNOT00000002418
|
Muc20
|
mucin 20, cell surface associated |
| chr10_+_58860940 | 2.69 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr1_+_61522298 | 2.64 |
ENSRNOT00000029111
|
Zfp51
|
zinc finger protein 51 |
| chr2_+_125752130 | 2.60 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr13_-_89242443 | 2.59 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chr10_+_95242986 | 2.57 |
ENSRNOT00000065589
ENSRNOT00000080460 |
RGD1565033
|
similar to hypothetical protein LOC284018 isoform b |
| chr10_-_59049482 | 2.56 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr2_-_26011429 | 2.55 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr8_-_21523540 | 2.50 |
ENSRNOT00000085060
|
Zfp266
|
zinc finger protein 266 |
| chr7_+_101069104 | 2.47 |
ENSRNOT00000056894
|
AABR07058124.2
|
|
| chr9_-_89193821 | 2.44 |
ENSRNOT00000090881
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr12_+_42097626 | 2.42 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr3_+_138597638 | 2.36 |
ENSRNOT00000031623
|
Zfp133
|
zinc finger protein 133 |
| chr6_+_137953545 | 2.34 |
ENSRNOT00000006804
|
Crip2
|
cysteine-rich protein 2 |
| chr10_+_37724915 | 2.29 |
ENSRNOT00000008477
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr20_+_13836030 | 2.19 |
ENSRNOT00000093157
ENSRNOT00000001659 |
Cabin1
|
calcineurin binding protein 1 |
| chr10_+_47282208 | 2.18 |
ENSRNOT00000057953
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr1_-_234749447 | 2.17 |
ENSRNOT00000016930
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr11_-_74315248 | 2.16 |
ENSRNOT00000002346
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr10_+_47281786 | 2.16 |
ENSRNOT00000089123
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr20_-_11815647 | 2.15 |
ENSRNOT00000001639
|
Itgb2
|
integrin subunit beta 2 |
| chr1_-_240601744 | 2.13 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr15_-_51400606 | 2.12 |
ENSRNOT00000023318
|
Chmp7
|
charged multivesicular body protein 7 |
| chr2_-_119007835 | 2.08 |
ENSRNOT00000087803
|
Gnb4
|
G protein subunit beta 4 |
| chr1_+_252894663 | 2.07 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr18_-_4294136 | 2.05 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr2_+_76923591 | 2.04 |
ENSRNOT00000042759
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chr6_+_127333590 | 2.04 |
ENSRNOT00000063970
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr8_-_102149912 | 2.02 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr19_-_955626 | 1.98 |
ENSRNOT00000017774
|
Bean1
|
brain expressed, associated with NEDD4, 1 |
| chr19_-_43528851 | 1.98 |
ENSRNOT00000036972
|
Mlkl
|
mixed lineage kinase domain like pseudokinase |
| chr10_-_56276764 | 1.97 |
ENSRNOT00000049048
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr9_-_61418679 | 1.96 |
ENSRNOT00000078800
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr1_-_73682247 | 1.96 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr3_+_92640752 | 1.96 |
ENSRNOT00000007604
|
Slc1a2
|
solute carrier family 1 member 2 |
| chr2_-_44379438 | 1.95 |
ENSRNOT00000073469
|
Il31ra
|
interleukin 31 receptor A |
| chr7_+_11356118 | 1.91 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr5_+_18901039 | 1.89 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr17_+_76532611 | 1.87 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_+_147661618 | 1.87 |
ENSRNOT00000011054
|
Bsdc1
|
BSD domain containing 1 |
| chr2_+_4942775 | 1.85 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr7_-_15073052 | 1.85 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr6_+_113898420 | 1.83 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr20_+_48881194 | 1.82 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr1_+_53874860 | 1.76 |
ENSRNOT00000090486
|
Tcte2
|
t-complex-associated testis expressed 2 |
| chr8_+_4440876 | 1.74 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr2_+_159138758 | 1.71 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chrX_+_40258493 | 1.71 |
ENSRNOT00000033010
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr1_+_64928503 | 1.68 |
ENSRNOT00000086274
|
Vom2r80
|
vomeronasal 2 receptor, 80 |
| chrX_-_159891326 | 1.68 |
ENSRNOT00000001154
|
Rbmx
|
RNA binding motif protein, X-linked |
| chr11_+_38727048 | 1.67 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr17_-_14687408 | 1.64 |
ENSRNOT00000088542
|
AC120310.1
|
|
| chr17_+_81798756 | 1.64 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr8_+_107875991 | 1.64 |
ENSRNOT00000090299
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_57816621 | 1.62 |
ENSRNOT00000090748
|
AABR07057150.1
|
|
| chr1_-_250626844 | 1.58 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr4_+_88271061 | 1.56 |
ENSRNOT00000087374
|
Vom1r86
|
vomeronasal 1 receptor 86 |
| chr1_-_60407295 | 1.55 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
| chr1_+_64849657 | 1.54 |
ENSRNOT00000081724
ENSRNOT00000086867 |
Vom2r12
|
vomeronasal 2 receptor, 12 |
| chr1_-_199341302 | 1.53 |
ENSRNOT00000073596
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr8_-_109576353 | 1.53 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr3_+_72134731 | 1.51 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr1_+_170578889 | 1.49 |
ENSRNOT00000025906
|
Ilk
|
integrin-linked kinase |
| chr2_-_80408672 | 1.47 |
ENSRNOT00000067638
|
Fam105a
|
family with sequence similarity 105, member A |
| chr15_+_105640097 | 1.46 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr1_-_216971183 | 1.46 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr2_-_45077219 | 1.45 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr7_+_37101391 | 1.44 |
ENSRNOT00000029764
|
Eea1
|
early endosome antigen 1 |
| chrX_-_83151511 | 1.44 |
ENSRNOT00000057378
|
Hdx
|
highly divergent homeobox |
| chr8_-_107952530 | 1.41 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr5_-_4972124 | 1.40 |
ENSRNOT00000081661
|
Xkr9
|
XK related 9 |
| chr11_+_34316295 | 1.39 |
ENSRNOT00000081834
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr1_-_31545559 | 1.39 |
ENSRNOT00000017779
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr18_+_62805410 | 1.39 |
ENSRNOT00000086679
|
Gnal
|
G protein subunit alpha L |
| chr1_-_212568224 | 1.38 |
ENSRNOT00000024976
|
LOC100911225
|
fucose mutarotase-like |
| chr5_+_64053946 | 1.37 |
ENSRNOT00000011622
|
Invs
|
inversin |
| chr12_+_19512591 | 1.35 |
ENSRNOT00000060024
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr1_-_142183884 | 1.33 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr12_-_19439977 | 1.33 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr5_-_2803855 | 1.32 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chrX_+_123350346 | 1.32 |
ENSRNOT00000017026
|
Slc25a43
|
solute carrier family 25, member 43 |
| chr12_-_30770791 | 1.31 |
ENSRNOT00000093734
|
Sfswap
|
splicing factor SWAP homolog |
| chr12_-_19440501 | 1.29 |
ENSRNOT00000060048
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr2_+_206342066 | 1.29 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr7_+_118502676 | 1.29 |
ENSRNOT00000005925
|
Commd5
|
COMM domain containing 5 |
| chr10_-_56850085 | 1.28 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr8_+_50200069 | 1.28 |
ENSRNOT00000023677
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr10_+_59529785 | 1.27 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr5_+_116420690 | 1.23 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr5_+_74649765 | 1.23 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chrX_-_20807216 | 1.21 |
ENSRNOT00000075757
|
Fam156b
|
family with sequence similarity 156, member B |
| chr2_+_196608496 | 1.20 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr2_-_188553289 | 1.19 |
ENSRNOT00000088822
|
Trim46
|
tripartite motif-containing 46 |
| chr14_-_114047527 | 1.19 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr1_+_202770775 | 1.19 |
ENSRNOT00000027704
|
Wdr11
|
WD repeat domain 11 |
| chr2_-_149325913 | 1.19 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chr12_+_19513100 | 1.18 |
ENSRNOT00000030161
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr10_-_56849255 | 1.13 |
ENSRNOT00000025491
|
RGD1308134
|
similar to RIKEN cDNA 1110020A23 |
| chr13_+_92146586 | 1.12 |
ENSRNOT00000004659
|
Olr1588
|
olfactory receptor 1588 |
| chr4_+_7258176 | 1.11 |
ENSRNOT00000061992
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr1_-_74638587 | 1.11 |
ENSRNOT00000046047
|
Vom2r30
|
vomeronasal 2 receptor, 30 |
| chr8_-_112807598 | 1.11 |
ENSRNOT00000015344
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr3_-_77525518 | 1.10 |
ENSRNOT00000049513
|
Olr663
|
olfactory receptor 663 |
| chr6_-_146470456 | 1.10 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr9_+_17122284 | 1.10 |
ENSRNOT00000077749
|
Polr1c
|
RNA polymerase I subunit C |
| chr5_-_2037038 | 1.09 |
ENSRNOT00000008680
|
Tmem70
|
transmembrane protein 70 |
| chr15_+_35504845 | 1.09 |
ENSRNOT00000075687
|
Olr1297
|
olfactory receptor 1297 |
| chr4_-_163445302 | 1.02 |
ENSRNOT00000087106
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr9_-_24451435 | 1.00 |
ENSRNOT00000060805
|
Defb49
|
defensin beta 49 |
| chr10_+_15696824 | 0.99 |
ENSRNOT00000079290
|
Il9r
|
interleukin 9 receptor |
| chr4_-_163445136 | 0.98 |
ENSRNOT00000080299
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr4_+_1566448 | 0.98 |
ENSRNOT00000088227
|
Olr1243
|
olfactory receptor 1243 |
| chr10_+_7041510 | 0.97 |
ENSRNOT00000003514
|
Carhsp1
|
calcium regulated heat stable protein 1 |
| chr2_+_225645568 | 0.93 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr8_-_43796224 | 0.90 |
ENSRNOT00000084315
|
Olr1330
|
olfactory receptor 1330 |
| chr9_+_98113346 | 0.88 |
ENSRNOT00000026912
|
Prlh
|
prolactin releasing hormone |
| chr7_-_130408187 | 0.88 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chr4_-_163423628 | 0.87 |
ENSRNOT00000079526
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr6_+_128738388 | 0.83 |
ENSRNOT00000050204
|
Rpl6-ps1
|
ribosomal protein L6, pseudogene 1 |
| chr13_-_101697684 | 0.82 |
ENSRNOT00000078834
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr17_+_44522140 | 0.79 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_-_73733788 | 0.77 |
ENSRNOT00000025338
|
Leng8
|
leukocyte receptor cluster member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.5 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 3.0 | 9.0 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 2.7 | 8.2 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 2.0 | 6.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 2.0 | 6.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.8 | 5.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 1.7 | 5.2 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.7 | 6.9 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.5 | 5.9 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 1.3 | 5.1 | GO:0051025 | microglial cell activation involved in immune response(GO:0002282) negative regulation of immunoglobulin secretion(GO:0051025) macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 1.2 | 3.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 3.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.1 | 3.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 1.1 | 3.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 1.0 | 4.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.9 | 4.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 3.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.9 | 3.5 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.9 | 2.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.9 | 4.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.8 | 5.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.7 | 3.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.7 | 2.2 | GO:0045608 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.7 | 4.8 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.7 | 3.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.7 | 3.3 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.6 | 1.9 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.6 | 3.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.6 | 1.7 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 0.6 | 7.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.6 | 1.7 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.5 | 1.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 9.2 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.5 | 4.6 | GO:0000050 | urea cycle(GO:0000050) |
| 0.5 | 1.5 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.5 | 3.0 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.5 | 2.4 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.5 | 1.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.4 | 1.3 | GO:0071613 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.4 | 2.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.4 | 2.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 4.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 1.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 2.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 1.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.3 | 6.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 4.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.3 | 1.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 0.9 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.3 | 6.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 4.6 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 8.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 0.8 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.3 | 2.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.7 | GO:0019541 | propionate metabolic process(GO:0019541) |
| 0.2 | 2.6 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 0.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 0.6 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 | 2.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 2.8 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.2 | 2.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 1.3 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.2 | 2.0 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 1.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 2.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 0.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.2 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 2.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 2.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.4 | GO:0002677 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of chronic inflammatory response(GO:0002677) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 4.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 3.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 3.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 3.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 4.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 3.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 3.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.1 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.1 | 3.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 4.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 1.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 2.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.0 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 1.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 3.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 2.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 4.6 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.6 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 3.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 2.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 2.0 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 5.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.6 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 1.4 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 3.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 3.6 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 6.9 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 1.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.8 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 3.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.6 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 2.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.5 | 4.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 1.3 | 6.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.2 | 6.1 | GO:0097413 | Lewy body(GO:0097413) |
| 1.1 | 3.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.9 | 3.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.6 | 3.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 1.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 1.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 3.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 2.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 3.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 1.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 6.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 17.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 3.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 2.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.3 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 1.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 6.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 12.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 4.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 4.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 5.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 10.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 9.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 3.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 10.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 52.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 6.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 15.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 2.5 | 10.1 | GO:0016160 | amylase activity(GO:0016160) |
| 2.2 | 6.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.0 | 5.9 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.8 | 5.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 1.6 | 4.9 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 1.6 | 8.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.5 | 4.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.3 | 26.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 1.2 | 6.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 1.2 | 4.8 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.1 | 3.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.1 | 4.2 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.1 | 3.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.9 | 7.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 6.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 3.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.7 | 3.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.7 | 2.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.7 | 4.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.6 | 9.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 4.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.5 | 1.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.4 | 2.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 1.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 2.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 2.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 3.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 4.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 4.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 5.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 1.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 7.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 3.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 2.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 6.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 3.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.3 | 4.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 11.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 2.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 3.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 2.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 4.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 3.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 0.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 2.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 3.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 8.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 4.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.1 | GO:0008009 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 3.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 6.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 3.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 1.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 2.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 3.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 3.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 4.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 1.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 14.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 2.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 6.9 | GO:0016491 | oxidoreductase activity(GO:0016491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 7.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 3.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 5.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.2 | 17.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 6.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 10.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 1.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 6.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.3 | 4.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 4.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.3 | 11.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 2.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 19.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 4.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 4.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 5.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 5.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 4.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 6.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 21.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 2.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |