Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
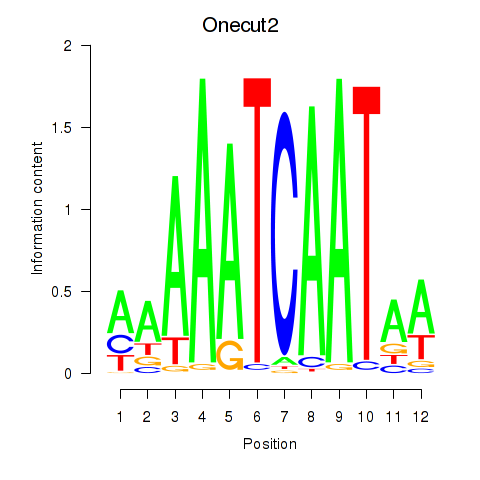
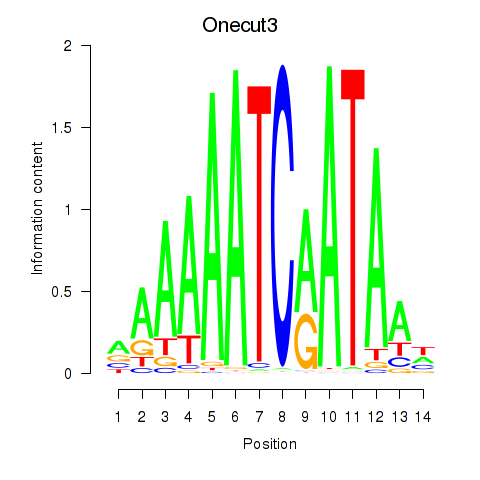
Results for Onecut2_Onecut3
Z-value: 0.82
Transcription factors associated with Onecut2_Onecut3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut2
|
ENSRNOG00000052665 | one cut domain, family member 2 |
|
Onecut3
|
ENSRNOG00000059889 | one cut domain, family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut2 | rn6_v1_chr18_+_59830363_59830422 | 0.44 | 9.8e-17 | Click! |
| Onecut3 | rn6_v1_chr7_-_12085226_12085226 | 0.09 | 1.2e-01 | Click! |
Activity profile of Onecut2_Onecut3 motif
Sorted Z-values of Onecut2_Onecut3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_161850875 | 46.32 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr2_+_60180215 | 25.66 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr1_-_258766881 | 25.60 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr10_+_65767053 | 23.95 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chr1_-_76614279 | 23.41 |
ENSRNOT00000041367
ENSRNOT00000089371 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr1_-_76780230 | 21.64 |
ENSRNOT00000002046
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr1_-_89560719 | 19.09 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr6_-_127620296 | 18.79 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr1_-_89560469 | 17.48 |
ENSRNOT00000079091
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr13_+_91080341 | 17.24 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr11_-_81660395 | 17.06 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr2_-_98610368 | 16.85 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr15_+_33478148 | 16.70 |
ENSRNOT00000089954
|
AC115371.2
|
|
| chr4_-_48928372 | 14.69 |
ENSRNOT00000083938
|
Tspan12
|
tetraspanin 12 |
| chrX_+_51286737 | 13.85 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr2_+_93827504 | 13.18 |
ENSRNOT00000032059
|
Pmp2
|
peripheral myelin protein 2 |
| chr5_-_127878792 | 12.91 |
ENSRNOT00000014436
|
Zyg11b
|
zyg-11 family member B, cell cycle regulator |
| chr17_+_72160735 | 12.83 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_-_76517134 | 11.96 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr1_+_248428099 | 11.71 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr7_+_3216497 | 11.38 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr2_+_235596907 | 11.14 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr16_+_18736154 | 11.05 |
ENSRNOT00000015723
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr9_-_4327679 | 10.50 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr2_+_241909832 | 10.47 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_145948996 | 10.42 |
ENSRNOT00000043476
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr3_+_129753742 | 9.84 |
ENSRNOT00000007998
ENSRNOT00000080581 |
Snap25
|
synaptosomal-associated protein 25 |
| chr9_-_85617954 | 9.76 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr15_-_14737704 | 9.64 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr7_+_14441476 | 9.47 |
ENSRNOT00000093687
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr13_+_89586283 | 9.38 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr11_+_80358211 | 9.37 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr10_+_35343189 | 9.23 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr7_+_70980422 | 9.14 |
ENSRNOT00000077912
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr16_-_74408030 | 8.94 |
ENSRNOT00000026418
|
Slc20a2
|
solute carrier family 20 member 2 |
| chrX_-_13279082 | 8.73 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr2_+_18392142 | 8.69 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_282557426 | 8.69 |
ENSRNOT00000088966
|
AABR07007134.1
|
|
| chr11_+_36588404 | 8.64 |
ENSRNOT00000083003
|
AC142178.3
|
|
| chr20_-_28263037 | 8.41 |
ENSRNOT00000030348
|
Edar
|
ectodysplasin-A receptor |
| chr3_+_129599353 | 8.33 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
| chr14_-_33977277 | 8.09 |
ENSRNOT00000002890
|
LOC100910308
|
multifunctional protein ADE2-like |
| chr9_+_6970507 | 7.80 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr8_-_70270309 | 7.66 |
ENSRNOT00000088311
|
AC107597.1
|
|
| chr3_+_5519990 | 7.63 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr1_+_256955652 | 7.63 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chrX_+_120901495 | 7.60 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr3_+_142383278 | 7.57 |
ENSRNOT00000017742
|
Foxa2
|
forkhead box A2 |
| chr2_-_231409496 | 7.44 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr10_-_27179254 | 7.43 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr5_-_105579959 | 7.38 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chrX_+_6273733 | 7.34 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr6_+_47940183 | 7.14 |
ENSRNOT00000011951
|
Adi1
|
acireductone dioxygenase 1 |
| chr15_-_23969011 | 7.05 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr17_-_4454701 | 7.04 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr16_+_23317953 | 7.03 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr1_-_175657485 | 6.91 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr10_+_53621375 | 6.89 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr3_+_11653529 | 6.85 |
ENSRNOT00000091048
|
Ak1
|
adenylate kinase 1 |
| chr5_+_27312928 | 6.85 |
ENSRNOT00000078102
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr1_+_219764001 | 6.65 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr5_+_76092287 | 6.61 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr3_+_51687809 | 6.50 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr2_+_115074344 | 6.36 |
ENSRNOT00000073657
|
LOC100910500
|
olfactory receptor 144-like |
| chr1_+_264741911 | 6.26 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr5_-_88612626 | 6.09 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr2_-_209993250 | 6.08 |
ENSRNOT00000024519
|
Prok1
|
prokineticin 1 |
| chr2_-_258932200 | 6.00 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr8_-_22821397 | 5.99 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr15_-_33752665 | 5.89 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr5_-_7941822 | 5.80 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_137860946 | 5.68 |
ENSRNOT00000031334
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr8_-_87245951 | 5.61 |
ENSRNOT00000015299
|
Tmem30a
|
transmembrane protein 30A |
| chr6_-_95934296 | 5.53 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr5_+_49311030 | 5.32 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr1_-_143392532 | 5.28 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr8_-_80631873 | 5.20 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr11_-_61470046 | 5.17 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr15_-_28746042 | 5.11 |
ENSRNOT00000017730
|
Sall2
|
spalt-like transcription factor 2 |
| chr19_+_14346197 | 5.11 |
ENSRNOT00000043480
|
Gm18025
|
predicted gene, 18025 |
| chrX_-_64908682 | 5.04 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr2_-_35550204 | 4.99 |
ENSRNOT00000084377
|
Olr1280
|
olfactory receptor 1280 |
| chr15_+_12407524 | 4.73 |
ENSRNOT00000009249
|
Psmd6
|
proteasome 26S subunit, non-ATPase 6 |
| chrM_+_2740 | 4.73 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chrX_+_44907521 | 4.68 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr18_-_85980833 | 4.67 |
ENSRNOT00000058179
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr11_-_15858281 | 4.56 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr1_-_63457134 | 4.51 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr1_-_263885169 | 4.50 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr14_-_103321270 | 4.48 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr1_+_193424812 | 4.46 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chr3_-_8924032 | 4.45 |
ENSRNOT00000023527
ENSRNOT00000085042 |
Sh3glb2
|
SH3 domain-containing GRB2-like endophilin B2 |
| chr11_-_76888178 | 4.45 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr4_-_160334910 | 4.33 |
ENSRNOT00000086772
|
Prmt8
|
protein arginine methyltransferase 8 |
| chr10_-_91986632 | 4.29 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr8_+_14811627 | 4.18 |
ENSRNOT00000043392
|
LOC689840
|
LRRGT00142 |
| chr7_+_93376277 | 4.15 |
ENSRNOT00000017602
|
LOC100359763
|
ribosomal protein S25-like |
| chr16_+_74408183 | 4.12 |
ENSRNOT00000036506
|
Smim19
|
small integral membrane protein 19 |
| chrX_-_100632562 | 4.10 |
ENSRNOT00000073416
|
LOC102555578
|
zinc finger protein 120-like |
| chr8_+_97535777 | 4.07 |
ENSRNOT00000068394
|
Adamts7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr11_+_86421106 | 3.99 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr14_-_22009300 | 3.98 |
ENSRNOT00000088491
ENSRNOT00000087923 |
Csn1s1
|
casein alpha s1 |
| chr8_+_100260049 | 3.95 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr3_-_143655864 | 3.95 |
ENSRNOT00000088666
|
AABR07054167.1
|
|
| chr11_+_75434197 | 3.88 |
ENSRNOT00000032569
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr10_+_80790168 | 3.87 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr17_-_43537293 | 3.76 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chrX_-_82986051 | 3.68 |
ENSRNOT00000077587
|
Hdx
|
highly divergent homeobox |
| chr2_+_113007549 | 3.65 |
ENSRNOT00000017758
|
Tnfsf10
|
tumor necrosis factor superfamily member 10 |
| chr3_-_83306781 | 3.63 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr7_-_106465830 | 3.60 |
ENSRNOT00000044610
|
AABR07058253.1
|
|
| chr10_-_66848388 | 3.59 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr7_-_117788550 | 3.58 |
ENSRNOT00000021775
|
MGC94207
|
similar to RIKEN cDNA C030006K11 |
| chr10_-_15465404 | 3.50 |
ENSRNOT00000077826
ENSRNOT00000027593 |
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr6_+_27535020 | 3.45 |
ENSRNOT00000076512
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr15_-_27408258 | 3.43 |
ENSRNOT00000043588
|
RGD1562558
|
similar to Cytochrome c, somatic |
| chrX_+_74304292 | 3.42 |
ENSRNOT00000057623
|
LOC680227
|
LRRGT00193 |
| chr9_+_66045962 | 3.28 |
ENSRNOT00000058491
|
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr14_+_1937041 | 3.27 |
ENSRNOT00000000040
|
Tmed11
|
transmembrane emp24 protein transport domain containing 11 |
| chr3_-_143778353 | 3.25 |
ENSRNOT00000050369
|
P22k15
|
cystatin related protein 2 |
| chr8_-_43480777 | 3.23 |
ENSRNOT00000060074
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr19_+_14345993 | 3.22 |
ENSRNOT00000084913
|
Gm18025
|
predicted gene, 18025 |
| chr20_+_6356423 | 3.22 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr17_-_1999596 | 3.14 |
ENSRNOT00000072220
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr20_-_3439983 | 3.10 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr6_-_26138414 | 3.10 |
ENSRNOT00000034712
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chr1_+_172892134 | 3.08 |
ENSRNOT00000087918
|
Olr276
|
olfactory receptor 276 |
| chr2_-_96032722 | 3.06 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr4_-_79052633 | 3.05 |
ENSRNOT00000012694
|
RGD1563352
|
similar to ribosomal protein S11 |
| chr8_-_43534620 | 3.02 |
ENSRNOT00000035386
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr8_+_59900651 | 2.93 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr4_-_23119005 | 2.92 |
ENSRNOT00000048061
|
Steap4
|
STEAP4 metalloreductase |
| chr4_-_30380119 | 2.91 |
ENSRNOT00000036460
|
Pon2
|
paraoxonase 2 |
| chr11_+_40509078 | 2.88 |
ENSRNOT00000023108
|
AABR07033851.1
|
|
| chr10_+_59259955 | 2.80 |
ENSRNOT00000021816
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr7_+_8210182 | 2.79 |
ENSRNOT00000075032
|
Olr1049
|
olfactory receptor 1049 |
| chr2_-_242828629 | 2.78 |
ENSRNOT00000043369
|
LOC499715
|
LRRGT00095 |
| chr5_-_107039925 | 2.74 |
ENSRNOT00000049705
|
Ifna5
|
interferon, alpha 5 |
| chrX_-_45284341 | 2.73 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr9_-_50762082 | 2.73 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr1_-_103811148 | 2.72 |
ENSRNOT00000030162
|
Mrgprx2l
|
MAS-related G protein-coupled receptor, member X2-like |
| chr3_-_77883067 | 2.71 |
ENSRNOT00000087979
|
Olr677
|
olfactory receptor 677 |
| chrX_-_84821775 | 2.70 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr3_-_158328881 | 2.68 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr7_-_12326392 | 2.68 |
ENSRNOT00000039728
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chrX_-_81817101 | 2.67 |
ENSRNOT00000003730
|
AABR07039648.1
|
|
| chr15_+_35571103 | 2.66 |
ENSRNOT00000085376
|
Olr1293
|
olfactory receptor 1293 |
| chr2_-_111793326 | 2.65 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chrX_+_45213228 | 2.64 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chrX_-_124576133 | 2.59 |
ENSRNOT00000041904
|
Rhox3
|
reproductive homeobox on X chromosome 3 |
| chr5_+_187312 | 2.55 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr9_-_17212628 | 2.54 |
ENSRNOT00000002656
ENSRNOT00000007876 |
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr8_-_19333047 | 2.50 |
ENSRNOT00000048500
|
Olr1144
|
olfactory receptor 1144 |
| chr18_-_52047816 | 2.44 |
ENSRNOT00000018368
ENSRNOT00000077491 |
LOC100174910
|
glutaredoxin-like protein |
| chr18_-_74059533 | 2.44 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr15_-_27798408 | 2.43 |
ENSRNOT00000088981
|
Tep1
|
telomerase associated protein 1 |
| chrX_-_83864150 | 2.42 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr15_+_51303909 | 2.40 |
ENSRNOT00000085237
ENSRNOT00000058663 |
Loxl2
|
lysyl oxidase-like 2 |
| chr8_+_76754492 | 2.35 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr3_-_73484676 | 2.35 |
ENSRNOT00000040041
|
Olr481
|
olfactory receptor 481 |
| chrX_-_74968405 | 2.34 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr19_-_39670909 | 2.31 |
ENSRNOT00000078388
|
AABR07043776.1
|
|
| chr12_-_38916237 | 2.30 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr17_+_53962444 | 2.26 |
ENSRNOT00000080101
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr7_-_17859653 | 2.26 |
ENSRNOT00000030164
|
Zscan4f
|
zinc finger and SCAN domain containing 4F |
| chrX_-_31013030 | 2.25 |
ENSRNOT00000004451
|
AABR07037798.1
|
|
| chr4_-_118342176 | 2.25 |
ENSRNOT00000032477
|
AC139642.2
|
|
| chr4_+_78140483 | 2.22 |
ENSRNOT00000048576
|
Zfp862
|
zinc finger protein 862 |
| chr9_+_40817654 | 2.22 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr3_+_145882819 | 2.21 |
ENSRNOT00000042110
|
LOC102549291
|
zinc finger protein 120-like |
| chr2_-_149088787 | 2.20 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr13_+_82355886 | 2.19 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr8_+_19810959 | 2.18 |
ENSRNOT00000049712
|
Olr1156
|
olfactory receptor 1156 |
| chr1_+_264059374 | 2.14 |
ENSRNOT00000075397
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr3_+_134440195 | 2.13 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr3_-_52849907 | 2.12 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr10_-_62393673 | 2.11 |
ENSRNOT00000070956
|
LOC100909578
|
olfactory receptor 1496-like |
| chr1_+_91746486 | 2.10 |
ENSRNOT00000047772
|
AC136661.1
|
|
| chr7_+_144565392 | 2.08 |
ENSRNOT00000021589
|
Hoxc11
|
homeobox C11 |
| chr20_-_31402483 | 2.08 |
ENSRNOT00000033529
|
H2afy2
|
H2A histone family, member Y2 |
| chr1_+_84584596 | 2.07 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chr16_-_16762240 | 2.06 |
ENSRNOT00000080746
|
AABR07024790.1
|
|
| chr2_+_115125790 | 2.06 |
ENSRNOT00000074937
|
LOC100909862
|
olfactory receptor 144-like |
| chr15_+_35942770 | 2.05 |
ENSRNOT00000072814
|
Olr1278
|
olfactory receptor 1278 |
| chr13_+_56096834 | 2.04 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr14_-_15709518 | 2.03 |
ENSRNOT00000051054
|
RGD1559999
|
similar to DNA segment, Chr 5, ERATO Doi 577, expressed |
| chr4_-_120414118 | 2.01 |
ENSRNOT00000072795
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr3_+_159902441 | 2.00 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr1_-_150262213 | 2.00 |
ENSRNOT00000049595
|
Olr32
|
olfactory receptor 32 |
| chr3_+_158851436 | 1.99 |
ENSRNOT00000075251
|
LOC100912432
|
olfactory receptor 8B8-like |
| chr5_-_107416695 | 1.99 |
ENSRNOT00000045433
|
Ifna2
|
interferon alpha 2 |
| chr6_-_41870046 | 1.98 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr1_+_84685931 | 1.98 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr4_-_129515435 | 1.98 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr4_+_64088900 | 1.96 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr4_+_1371401 | 1.95 |
ENSRNOT00000071676
|
LOC100910899
|
olfactory receptor 143-like |
| chr1_-_90208036 | 1.94 |
ENSRNOT00000076255
ENSRNOT00000076967 |
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr4_+_121778227 | 1.92 |
ENSRNOT00000084689
|
Vom1r93
|
vomeronasal 1 receptor 93 |
| chr5_-_33664435 | 1.91 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_-_11400805 | 1.90 |
ENSRNOT00000027634
|
Dapk3
|
death-associated protein kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut2_Onecut3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 36.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 6.3 | 18.8 | GO:0033986 | response to methanol(GO:0033986) |
| 6.1 | 18.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 4.6 | 13.8 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 2.9 | 11.7 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 2.9 | 17.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.9 | 25.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 2.6 | 10.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 2.5 | 7.6 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 2.4 | 9.8 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.4 | 7.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 2.3 | 6.8 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 2.2 | 24.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.0 | 6.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.9 | 7.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.8 | 5.5 | GO:2000729 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.8 | 5.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.7 | 6.7 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.5 | 4.5 | GO:0010034 | response to acetate(GO:0010034) |
| 1.5 | 4.4 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 1.4 | 8.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.3 | 8.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 1.3 | 9.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.3 | 10.4 | GO:0048840 | otolith development(GO:0048840) |
| 1.3 | 11.4 | GO:0001554 | luteolysis(GO:0001554) |
| 1.2 | 7.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.1 | 9.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.1 | 4.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 1.0 | 11.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 1.0 | 27.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.0 | 10.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.9 | 5.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.9 | 6.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.9 | 11.1 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.8 | 46.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.8 | 2.3 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.7 | 4.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.7 | 2.7 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.6 | 4.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 7.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.6 | 2.8 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.6 | 1.7 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 2.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 7.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 2.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.5 | 2.0 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.5 | 5.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 2.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 5.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 3.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.4 | 15.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.4 | 1.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.4 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 12.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.4 | 4.4 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.4 | 10.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 2.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.4 | 11.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.4 | 2.0 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 | 1.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 2.9 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 1.4 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.4 | 7.8 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 1.8 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 3.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 6.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.3 | 9.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 3.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 0.9 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.3 | 2.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 | 1.2 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.3 | 2.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 7.6 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.3 | 1.6 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 3.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 12.9 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.2 | 2.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 1.6 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 1.1 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.2 | 4.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 2.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 4.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 2.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 2.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 1.8 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) hormone catabolic process(GO:0042447) |
| 0.2 | 1.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 1.9 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 1.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 7.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 5.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.2 | 2.0 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 6.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.9 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.2 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 4.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 2.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.6 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.5 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 2.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 1.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.9 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 1.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 0.7 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 1.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 2.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.3 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 0.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 11.7 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 1.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 88.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.1 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.7 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 7.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 8.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 1.0 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 1.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 2.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.0 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.5 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.8 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 4.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 3.5 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 24.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 3.6 | 18.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 2.3 | 45.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.7 | 10.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.5 | 4.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 1.1 | 3.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.9 | 12.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.9 | 10.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.8 | 9.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 13.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 2.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 5.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 1.5 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.5 | 2.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.5 | 6.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 9.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 7.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 4.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 2.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 2.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 22.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 2.8 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 1.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 7.4 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 5.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 5.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 4.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.2 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 12.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 7.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 4.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 164.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 15.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 9.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 5.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 3.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 2.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 32.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 15.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 7.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 12.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 6.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 13.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 5.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 5.5 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 44.3 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 46.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 8.6 | 25.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 7.3 | 36.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.6 | 10.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.6 | 10.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 2.5 | 17.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.3 | 11.7 | GO:0005534 | galactose binding(GO:0005534) |
| 2.2 | 6.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.9 | 7.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.8 | 8.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.5 | 35.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.5 | 10.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.5 | 4.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.4 | 17.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.3 | 5.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.1 | 3.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.1 | 7.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.0 | 34.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.0 | 7.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.0 | 2.9 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.9 | 9.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.9 | 27.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 3.5 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.9 | 4.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.9 | 14.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.9 | 9.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 13.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.7 | 2.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.6 | 6.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 1.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 8.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.6 | 1.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 2.1 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.5 | 7.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 2.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 2.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.5 | 2.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 4.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 3.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.4 | 9.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 1.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.4 | 2.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 6.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.4 | 43.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 8.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 4.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 2.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 0.9 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.3 | 11.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 6.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 1.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.3 | 7.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 2.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 5.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 1.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 8.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 6.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 5.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.7 | GO:0045295 | alpha-catenin binding(GO:0045294) gamma-catenin binding(GO:0045295) delta-catenin binding(GO:0070097) |
| 0.2 | 5.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 2.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 7.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 7.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 2.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 6.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 7.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 60.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 1.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 16.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 3.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 1.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 9.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 13.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 11.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 4.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 7.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 5.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 5.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 30.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 7.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 9.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 12.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 2.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 24.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.5 | 19.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 27.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.5 | 26.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.5 | 8.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.4 | 94.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 17.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 3.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 7.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 10.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 8.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 10.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 18.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 29.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.3 | 18.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.3 | 45.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.2 | 25.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.7 | 9.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.6 | 7.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.6 | 7.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.6 | 9.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.5 | 20.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 7.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 9.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 7.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 9.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 10.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 4.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 6.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 24.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 4.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 4.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 10.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 10.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 18.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 6.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 11.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 4.7 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 3.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 15.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 7.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.9 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |