Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
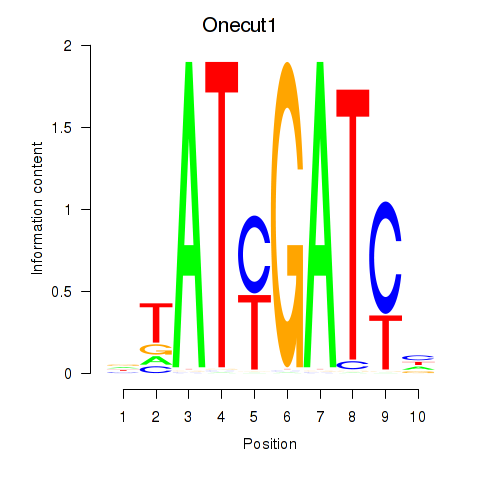
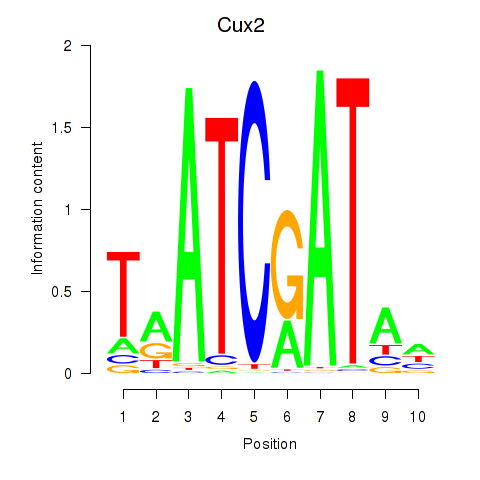
Results for Onecut1_Cux2
Z-value: 1.61
Transcription factors associated with Onecut1_Cux2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut1
|
ENSRNOG00000008095 | one cut homeobox 1 |
|
Cux2
|
ENSRNOG00000001259 | cut-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut1 | rn6_v1_chr8_+_81766041_81766041 | 0.83 | 7.3e-81 | Click! |
| Cux2 | rn6_v1_chr12_+_40018937_40018937 | 0.19 | 4.9e-04 | Click! |
Activity profile of Onecut1_Cux2 motif
Sorted Z-values of Onecut1_Cux2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_81639872 | 227.57 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chr5_-_78985990 | 210.53 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr5_-_124403195 | 205.42 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr13_+_91080341 | 187.01 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr2_+_114413410 | 178.87 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr17_+_9736577 | 105.82 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr17_+_9736786 | 99.84 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr10_+_65767053 | 85.92 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chr7_-_119689938 | 84.81 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr9_+_95501778 | 78.22 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr1_+_83714347 | 73.09 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr19_+_23389375 | 66.70 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr10_+_96639924 | 61.82 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr10_+_89285855 | 61.30 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr18_-_35071619 | 58.02 |
ENSRNOT00000075695
|
LOC100911558
|
serine protease inhibitor Kazal-type 3-like |
| chr10_+_89286047 | 57.23 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr18_-_59819113 | 49.17 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr7_-_34406318 | 46.01 |
ENSRNOT00000007331
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr3_+_142383278 | 41.50 |
ENSRNOT00000017742
|
Foxa2
|
forkhead box A2 |
| chr3_-_72171078 | 36.97 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr1_-_137359072 | 33.69 |
ENSRNOT00000014553
|
Klhl25
|
kelch-like family member 25 |
| chr8_+_117068582 | 32.23 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr7_-_102298522 | 31.83 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr2_-_98610368 | 29.51 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr5_+_136683592 | 28.48 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr4_-_44136815 | 27.99 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr2_-_258932200 | 27.29 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_-_235177275 | 26.68 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr2_+_68820615 | 25.45 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr2_+_22909569 | 23.56 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr2_-_178297172 | 19.62 |
ENSRNOT00000038543
|
Fnip2
|
folliculin interacting protein 2 |
| chr2_-_252451999 | 18.56 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr19_+_60017746 | 16.01 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr2_+_22910236 | 15.67 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr8_-_122904913 | 15.60 |
ENSRNOT00000015188
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr9_+_112360419 | 13.13 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr18_-_77579969 | 12.33 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr1_-_255815733 | 11.24 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr4_-_50312608 | 10.56 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr14_-_64476796 | 10.29 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chr10_+_80790168 | 9.80 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr6_-_108415093 | 9.48 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr8_-_87245951 | 9.36 |
ENSRNOT00000015299
|
Tmem30a
|
transmembrane protein 30A |
| chr18_-_52047816 | 8.98 |
ENSRNOT00000018368
ENSRNOT00000077491 |
LOC100174910
|
glutaredoxin-like protein |
| chr9_+_41096835 | 8.32 |
ENSRNOT00000033700
|
Amer3
|
APC membrane recruitment protein 3 |
| chr6_-_72786830 | 7.65 |
ENSRNOT00000009020
|
Dtd2
|
D-tyrosyl-tRNA deacylase 2 |
| chr1_-_170034892 | 6.84 |
ENSRNOT00000042936
|
Olr194
|
olfactory receptor 194 |
| chr3_+_307204 | 6.68 |
ENSRNOT00000062080
|
Nxph2
|
neurexophilin 2 |
| chr18_+_38847632 | 6.25 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr1_+_249574954 | 6.17 |
ENSRNOT00000074100
|
Cstf2t
|
cleavage stimulation factor subunit 2, tau variant |
| chr5_-_28164326 | 6.08 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr15_+_44799334 | 5.51 |
ENSRNOT00000018599
|
Nefl
|
neurofilament light |
| chr1_-_264975132 | 5.46 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr8_-_43192910 | 5.23 |
ENSRNOT00000060101
|
Olr1302
|
olfactory receptor 1302 |
| chrX_-_72370044 | 4.98 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr2_+_46186105 | 4.90 |
ENSRNOT00000071256
|
LOC100910378
|
olfactory receptor 145-like |
| chr8_-_1450138 | 4.38 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr3_-_165537940 | 4.10 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr5_-_62621737 | 4.07 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr1_-_167732783 | 3.96 |
ENSRNOT00000024965
|
Olr44
|
olfactory receptor 44 |
| chrX_-_113474164 | 2.91 |
ENSRNOT00000046804
|
Gucy2f
|
guanylate cyclase 2F |
| chr8_-_43055175 | 2.77 |
ENSRNOT00000074413
|
LOC103693054
|
olfactory receptor 8G1-like |
| chr2_+_179952227 | 2.65 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr15_+_35909279 | 2.62 |
ENSRNOT00000071711
|
Olr1279
|
olfactory receptor 1279 |
| chr13_+_91768256 | 2.50 |
ENSRNOT00000071206
|
LOC681470
|
similar to olfactory receptor 1403 |
| chr1_+_169944609 | 2.47 |
ENSRNOT00000023293
|
Olr188
|
olfactory receptor 188 |
| chr4_-_170912629 | 2.40 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr3_+_73249710 | 2.33 |
ENSRNOT00000083865
|
Olr464
|
olfactory receptor 464 |
| chr14_-_11546314 | 2.25 |
ENSRNOT00000090334
|
AABR07014355.1
|
|
| chrX_-_15467875 | 2.09 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr15_-_33752665 | 2.05 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr1_-_247985497 | 1.87 |
ENSRNOT00000078998
|
Ranbp6
|
RAN binding protein 6 |
| chr1_+_169698323 | 1.86 |
ENSRNOT00000075652
|
Olr163
|
olfactory receptor 163 |
| chr2_-_96032722 | 1.82 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr3_+_76035198 | 1.82 |
ENSRNOT00000080503
|
Olr596
|
olfactory receptor 596 |
| chrX_+_124894466 | 1.58 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr14_-_34218961 | 1.53 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr8_-_40564556 | 1.53 |
ENSRNOT00000075779
|
Olfr883
|
olfactory receptor 883 |
| chr1_+_214375515 | 1.28 |
ENSRNOT00000024863
|
Taldo1
|
transaldolase 1 |
| chr8_-_43908914 | 1.13 |
ENSRNOT00000072247
|
Olr1338
|
olfactory receptor 1338 |
| chr9_-_5330815 | 1.10 |
ENSRNOT00000014548
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr4_-_10269748 | 1.07 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr3_-_103111795 | 0.82 |
ENSRNOT00000006622
|
Olr778
|
olfactory receptor 778 |
| chrX_-_138148967 | 0.80 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr3_-_74256067 | 0.65 |
ENSRNOT00000047747
|
Olr522
|
olfactory receptor 522 |
| chrX_-_37353156 | 0.45 |
ENSRNOT00000086120
|
AABR07038021.1
|
|
| chrM_-_14061 | 0.38 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr8_+_42017493 | 0.35 |
ENSRNOT00000072062
|
LOC100911845
|
olfactory receptor 143-like |
| chr3_-_76626605 | 0.33 |
ENSRNOT00000051292
|
Olr625
|
olfactory receptor 625 |
| chr5_+_57845819 | 0.32 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chr2_+_143475323 | 0.08 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr3_-_81304181 | 0.06 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut1_Cux2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 75.9 | 227.6 | GO:0097037 | heme export(GO:0097037) |
| 68.6 | 205.7 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 35.8 | 178.9 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 31.2 | 187.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 22.2 | 66.7 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 21.1 | 210.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 15.5 | 61.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 15.3 | 46.0 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 13.8 | 41.5 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 12.2 | 73.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 11.9 | 118.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 8.1 | 32.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 7.8 | 85.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 7.7 | 84.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 5.9 | 194.5 | GO:0019835 | cytolysis(GO:0019835) |
| 3.4 | 37.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 3.2 | 25.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 3.1 | 58.0 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 2.8 | 11.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 2.7 | 16.0 | GO:0003383 | apical constriction(GO:0003383) |
| 2.6 | 10.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.6 | 39.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 2.6 | 28.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 2.6 | 10.3 | GO:1903015 | cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.8 | 5.5 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.7 | 15.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.7 | 5.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.2 | 13.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 1.2 | 18.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.9 | 4.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 9.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.8 | 6.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 12.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.7 | 78.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.6 | 5.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.5 | 32.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.4 | 1.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 6.1 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.3 | 7.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 1.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 1.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 2.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 1.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.2 | 4.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 14.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 19.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 2.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 4.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 8.3 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 2.9 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 24.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 56.9 | 227.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 29.3 | 205.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 14.3 | 85.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 4.8 | 61.8 | GO:0042627 | chylomicron(GO:0042627) |
| 2.9 | 66.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 2.7 | 16.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.2 | 269.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.9 | 178.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 1.8 | 5.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.5 | 170.2 | GO:0030175 | filopodium(GO:0030175) |
| 1.4 | 4.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 118.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.6 | 11.2 | GO:1990124 | CCR4-NOT complex(GO:0030014) messenger ribonucleoprotein complex(GO:1990124) |
| 0.6 | 23.0 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 33.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 6.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 13.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 339.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 23.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 9.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 15.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 21.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 7.7 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 42.1 | 210.5 | GO:0019862 | IgA binding(GO:0019862) |
| 26.7 | 187.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 23.7 | 118.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 20.6 | 61.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 20.2 | 242.4 | GO:0001848 | complement binding(GO:0001848) |
| 19.9 | 178.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 10.3 | 227.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 7.8 | 39.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 6.6 | 46.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 3.7 | 85.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 3.4 | 10.3 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 2.8 | 19.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.6 | 73.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.2 | 28.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 2.0 | 68.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 1.5 | 25.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.3 | 15.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.3 | 290.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 1.2 | 18.6 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 1.0 | 11.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 1.0 | 4.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.0 | 13.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.5 | 58.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 7.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 41.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.4 | 16.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 6.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 5.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 1.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 4.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 27.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 9.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 32.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 1.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 10.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 4.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 13.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.3 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 338.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 4.1 | 85.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 3.1 | 187.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 2.9 | 680.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.3 | 25.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.8 | 31.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.6 | 16.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 11.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 187.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 11.6 | 242.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 10.5 | 178.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 8.6 | 205.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 2.9 | 118.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 2.6 | 253.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 1.8 | 41.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.2 | 28.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.1 | 85.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.9 | 16.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.6 | 13.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.4 | 5.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 4.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 2.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 9.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 46.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 4.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 5.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |