Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
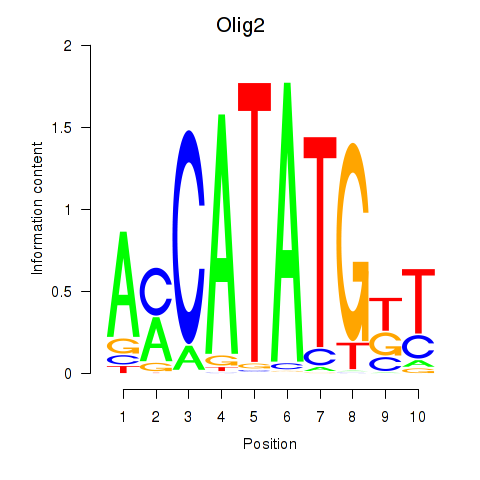
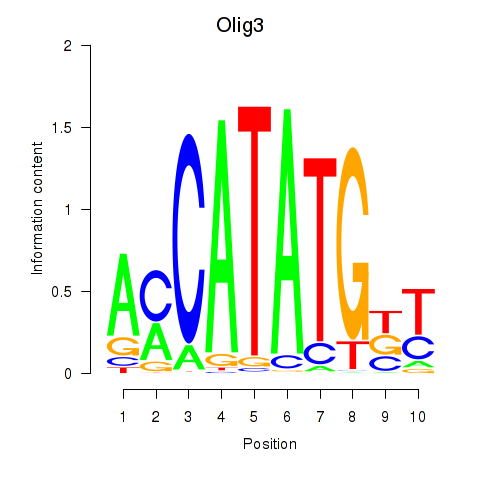
Results for Olig2_Olig3
Z-value: 0.74
Transcription factors associated with Olig2_Olig3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Olig2
|
ENSRNOG00000028658 | oligodendrocyte lineage transcription factor 2 |
|
Olig3
|
ENSRNOG00000012057 | oligodendrocyte transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Olig2 | rn6_v1_chr11_+_31389514_31389514 | -0.16 | 3.8e-03 | Click! |
| Olig3 | rn6_v1_chr1_+_14797766_14797766 | -0.10 | 7.9e-02 | Click! |
Activity profile of Olig2_Olig3 motif
Sorted Z-values of Olig2_Olig3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_139486775 | 18.15 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr1_-_227175096 | 16.96 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr20_-_5212624 | 13.23 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr4_-_103024841 | 12.95 |
ENSRNOT00000090283
|
AABR07061036.1
|
|
| chr6_+_139486942 | 12.64 |
ENSRNOT00000080320
|
AABR07065699.3
|
|
| chr10_+_19366793 | 12.54 |
ENSRNOT00000050610
|
Fam196b
|
family with sequence similarity 196, member B |
| chr4_-_183417667 | 12.53 |
ENSRNOT00000089160
|
Fam60a
|
family with sequence similarity 60, member A |
| chr6_-_44363915 | 12.38 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr8_+_5768811 | 12.38 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr3_+_20007192 | 12.37 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr4_-_155923079 | 12.12 |
ENSRNOT00000013308
|
Clec4a3
|
C-type lectin domain family 4, member A3 |
| chr1_-_198128857 | 11.43 |
ENSRNOT00000026496
|
Coro1a
|
coronin 1A |
| chr2_-_41871858 | 11.19 |
ENSRNOT00000039720
|
Gapt
|
Grb2-binding adaptor protein, transmembrane |
| chr2_-_54777729 | 11.07 |
ENSRNOT00000082548
|
C7
|
complement C7 |
| chr4_-_103258134 | 10.87 |
ENSRNOT00000086827
|
AABR07061052.1
|
|
| chrX_-_23187341 | 10.73 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr5_+_59783890 | 10.66 |
ENSRNOT00000066277
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr1_-_80744831 | 10.64 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr6_-_141291347 | 10.45 |
ENSRNOT00000008333
|
AABR07065789.1
|
|
| chr9_+_67763897 | 10.01 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chrX_+_105147534 | 9.94 |
ENSRNOT00000046288
|
Cenpi
|
centromere protein I |
| chr8_+_72743426 | 9.82 |
ENSRNOT00000072573
|
Rps27l
|
ribosomal protein S27-like |
| chr7_-_18793289 | 9.82 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr6_-_51297712 | 9.75 |
ENSRNOT00000082985
|
Prkar2b
|
protein kinase cAMP-dependent type 2 regulatory subunit beta |
| chrX_-_138435391 | 9.65 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr4_-_165192647 | 9.59 |
ENSRNOT00000086461
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr1_+_219403970 | 9.51 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr10_-_19574094 | 9.50 |
ENSRNOT00000059810
|
Dock2
|
dedicator of cytokinesis 2 |
| chrX_-_38026774 | 9.43 |
ENSRNOT00000074898
|
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr4_+_78378144 | 9.36 |
ENSRNOT00000059156
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr15_+_32828165 | 9.23 |
ENSRNOT00000060253
|
AABR07017902.1
|
|
| chr10_-_90312386 | 9.06 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr4_+_78378313 | 9.06 |
ENSRNOT00000083891
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr8_+_132869712 | 8.92 |
ENSRNOT00000008294
|
Cxcr6
|
C-X-C motif chemokine receptor 6 |
| chr9_-_53118613 | 8.89 |
ENSRNOT00000065581
|
Ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr2_+_193866951 | 8.72 |
ENSRNOT00000013393
|
S100a11
|
S100 calcium binding protein A11 |
| chr15_+_5772679 | 8.71 |
ENSRNOT00000043553
|
AABR07016992.1
|
|
| chr1_+_64074231 | 8.67 |
ENSRNOT00000077327
|
Lilrb3l
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3-like |
| chr3_-_148057523 | 8.49 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr1_+_63759638 | 8.32 |
ENSRNOT00000080799
ENSRNOT00000087669 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr4_+_96562725 | 8.32 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr2_+_122368265 | 8.24 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr9_+_12740885 | 8.14 |
ENSRNOT00000015210
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr4_-_103569159 | 8.00 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr20_+_3146856 | 8.00 |
ENSRNOT00000050159
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr10_-_70871066 | 7.95 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr2_-_105047984 | 7.73 |
ENSRNOT00000014970
|
Cpa3
|
carboxypeptidase A3 |
| chr1_-_197821936 | 7.69 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr8_-_49280901 | 7.61 |
ENSRNOT00000021390
|
Cd3g
|
CD3g molecule |
| chr7_+_3173287 | 7.58 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr19_-_29400355 | 7.57 |
ENSRNOT00000072100
|
LOC100909612
|
mitochondrial brown fat uncoupling protein 1-like |
| chr17_-_86657473 | 7.20 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr20_+_6211420 | 7.17 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr15_-_100348760 | 7.15 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr17_-_43815183 | 7.03 |
ENSRNOT00000073188
|
Hist1h2ail1
|
histone cluster 1, H2ai-like1 |
| chr1_-_61872975 | 7.02 |
ENSRNOT00000078809
|
AABR07001910.1
|
|
| chrX_+_77143892 | 6.97 |
ENSRNOT00000085227
|
Atp7a
|
ATPase copper transporting alpha |
| chr1_+_227592635 | 6.90 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr11_-_60819249 | 6.81 |
ENSRNOT00000043917
ENSRNOT00000042447 |
Cd200r1l
|
CD200 receptor 1-like |
| chr17_-_44643362 | 6.66 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr11_-_73678984 | 6.64 |
ENSRNOT00000002355
|
Fam43a
|
family with sequence similarity 43, member A |
| chr4_-_164049599 | 6.61 |
ENSRNOT00000078267
|
AABR07062183.1
|
|
| chr3_-_91217491 | 6.53 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chr5_+_173447784 | 6.51 |
ENSRNOT00000027251
|
Tnfrsf4
|
TNF receptor superfamily member 4 |
| chr10_-_59112788 | 6.30 |
ENSRNOT00000041886
|
Spns3
|
SPNS sphingolipid transporter 3 |
| chr1_-_101123402 | 6.27 |
ENSRNOT00000027976
|
Rpl13a
|
|
| chr8_-_22929294 | 6.23 |
ENSRNOT00000015609
|
Rab3d
|
RAB3D, member RAS oncogene family |
| chr6_-_143131118 | 6.22 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr1_+_88885937 | 6.08 |
ENSRNOT00000029719
|
Nfkbid
|
NFKB inhibitor delta |
| chr4_+_103524324 | 6.07 |
ENSRNOT00000087659
|
AABR07061068.2
|
|
| chr14_-_44225713 | 6.05 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr3_+_20641664 | 6.04 |
ENSRNOT00000044699
|
AABR07051741.1
|
|
| chr14_+_86751157 | 5.96 |
ENSRNOT00000067997
|
LOC100360491
|
60S ribosomal protein L13-like |
| chr7_-_54778848 | 5.91 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr4_-_164536556 | 5.84 |
ENSRNOT00000087796
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr3_+_19772056 | 5.84 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr9_+_112360419 | 5.76 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr1_+_64740487 | 5.69 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr3_+_20303979 | 5.67 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr8_-_48634797 | 5.67 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr14_-_18853315 | 5.65 |
ENSRNOT00000003794
|
Ppbp
|
pro-platelet basic protein |
| chr16_-_75481115 | 5.61 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr10_+_73868943 | 5.57 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr14_+_81043454 | 5.51 |
ENSRNOT00000043609
|
AC114393.1
|
|
| chr15_+_32809069 | 5.49 |
ENSRNOT00000070848
|
AABR07017902.1
|
|
| chr1_+_98398660 | 5.43 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr4_+_102290338 | 5.42 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr13_+_27312498 | 5.40 |
ENSRNOT00000003466
|
Serpinb7
|
serpin family B member 7 |
| chr12_+_19714324 | 5.37 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr3_-_145810834 | 5.36 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr10_-_56530842 | 5.34 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr1_+_199555722 | 5.34 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr14_-_86117578 | 5.33 |
ENSRNOT00000019288
|
Pold2
|
DNA polymerase delta 2, accessory subunit |
| chr1_-_73619356 | 5.27 |
ENSRNOT00000074352
|
Lilrb3
|
leukocyte immunoglobulin like receptor B3 |
| chrX_-_105390580 | 5.27 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr6_-_140572023 | 5.27 |
ENSRNOT00000072338
|
AABR07065772.1
|
|
| chr15_-_34612432 | 5.24 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chrX_-_76708878 | 5.21 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr10_-_112565146 | 5.20 |
ENSRNOT00000056238
|
Rpl5l1
|
ribosomal protein L5-like 1 |
| chr4_-_81241152 | 5.19 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr6_-_140102325 | 5.18 |
ENSRNOT00000072238
|
AABR07065750.2
|
|
| chr7_+_60316045 | 5.14 |
ENSRNOT00000064594
|
Lyc2
|
lysozyme C type 2 |
| chr7_+_72599479 | 5.12 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chrX_-_153491837 | 5.12 |
ENSRNOT00000089027
|
LOC100360413
|
eukaryotic translation elongation factor 1 alpha 1-like |
| chr16_+_90325304 | 5.12 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr10_-_70337532 | 5.09 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr1_+_225163391 | 5.09 |
ENSRNOT00000027305
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr2_+_38230757 | 4.96 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr20_+_3990820 | 4.94 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr10_-_4910305 | 4.91 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr16_-_25192675 | 4.88 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_98493978 | 4.87 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chrX_+_120901495 | 4.86 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr5_+_144927759 | 4.81 |
ENSRNOT00000051387
|
AABR07049918.1
|
|
| chr9_-_54351339 | 4.80 |
ENSRNOT00000080522
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr3_-_112985318 | 4.79 |
ENSRNOT00000015556
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr12_+_25119355 | 4.78 |
ENSRNOT00000034629
|
Lat2
|
linker for activation of T cells family, member 2 |
| chr13_-_79899479 | 4.78 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr1_-_47502952 | 4.74 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr3_+_19274273 | 4.74 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chrX_+_35869538 | 4.73 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr1_-_136073483 | 4.72 |
ENSRNOT00000043854
ENSRNOT00000082564 ENSRNOT00000076340 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr6_+_26566494 | 4.69 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr4_+_102876038 | 4.68 |
ENSRNOT00000085205
|
AABR07061022.3
|
|
| chr18_-_61788859 | 4.66 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr10_+_107469377 | 4.66 |
ENSRNOT00000087404
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_+_63964155 | 4.63 |
ENSRNOT00000078512
|
AABR07002001.1
|
|
| chr3_+_19045214 | 4.62 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr3_-_171286413 | 4.60 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr4_-_102124609 | 4.58 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr20_+_32717564 | 4.52 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr2_+_143656793 | 4.51 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr16_-_71237118 | 4.46 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chrX_-_104814145 | 4.43 |
ENSRNOT00000004909
|
Sytl4
|
synaptotagmin-like 4 |
| chr2_+_87418517 | 4.41 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chrX_-_154918095 | 4.38 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr1_+_265761738 | 4.37 |
ENSRNOT00000024898
|
Hps6
|
Hermansky-Pudlak syndrome 6 |
| chr9_-_81586116 | 4.36 |
ENSRNOT00000089952
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr4_-_78342863 | 4.32 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr14_+_110676090 | 4.30 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr1_-_221321459 | 4.22 |
ENSRNOT00000028406
ENSRNOT00000084800 |
Pola2
|
DNA polymerase alpha 2, accessory subunit |
| chr10_+_95770154 | 4.22 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr4_-_163227242 | 4.21 |
ENSRNOT00000091552
ENSRNOT00000077793 |
Clec7a
|
C-type lectin domain family 7, member A |
| chrX_-_115496045 | 4.19 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr2_+_76923591 | 4.17 |
ENSRNOT00000042759
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chr8_+_2604962 | 4.15 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chrX_+_15378789 | 4.15 |
ENSRNOT00000029272
|
Gata1
|
GATA binding protein 1 |
| chr10_+_17261541 | 4.14 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr1_-_63684189 | 4.13 |
ENSRNOT00000085651
|
Lilrc2
|
leukocyte immunoglobulin-like receptor, subfamily C, member 2 |
| chr1_-_266142538 | 4.12 |
ENSRNOT00000026792
|
Actr1a
|
ARP1 actin-related protein 1 homolog A, centractin alpha |
| chr9_-_81586469 | 4.11 |
ENSRNOT00000019848
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr3_+_111826297 | 4.09 |
ENSRNOT00000081897
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr16_-_83438561 | 4.07 |
ENSRNOT00000057461
|
Col4a2
|
collagen type IV alpha 2 chain |
| chr2_-_149444548 | 4.05 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr2_+_196594303 | 4.05 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr10_-_19715832 | 4.03 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr9_-_84894599 | 3.98 |
ENSRNOT00000018423
|
Hdac1l
|
histone deacetylase 1-like |
| chr10_+_87759769 | 3.96 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr4_-_164357619 | 3.96 |
ENSRNOT00000078114
|
Ly49i5
|
Ly49 inhibitory receptor 5 |
| chr18_+_55666027 | 3.94 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr15_+_42489377 | 3.94 |
ENSRNOT00000064306
|
Pbk
|
PDZ binding kinase |
| chr6_+_139177200 | 3.94 |
ENSRNOT00000084131
|
AABR07065676.1
|
|
| chr10_+_84182118 | 3.92 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr7_-_15852930 | 3.92 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr7_+_74350479 | 3.92 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr1_+_219468675 | 3.88 |
ENSRNOT00000025342
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr6_+_34030620 | 3.88 |
ENSRNOT00000091651
|
Laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr9_+_61738471 | 3.87 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr6_+_145546595 | 3.86 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr19_-_39267928 | 3.86 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_252505971 | 3.85 |
ENSRNOT00000021984
|
LOC56764
|
dnaj-like protein |
| chr10_-_63952726 | 3.83 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr4_+_102147211 | 3.82 |
ENSRNOT00000083239
|
AABR07060980.1
|
|
| chr10_+_104932616 | 3.81 |
ENSRNOT00000075706
|
RGD1562667
|
similar to leukocyte mono-Ig-like receptor2 |
| chr13_+_49005405 | 3.76 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr1_+_199495298 | 3.75 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr14_-_25585222 | 3.73 |
ENSRNOT00000042106
|
RGD1565048
|
similar to 60S ribosomal protein L9 |
| chr14_-_21299068 | 3.71 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr1_+_81474553 | 3.71 |
ENSRNOT00000083493
|
Phldb3
|
pleckstrin homology-like domain, family B, member 3 |
| chr2_+_186685104 | 3.70 |
ENSRNOT00000057022
|
Cd5l
|
Cd5 molecule-like |
| chr5_-_59638385 | 3.70 |
ENSRNOT00000060203
ENSRNOT00000077367 |
Rnf38
|
ring finger protein 38 |
| chr10_+_59725398 | 3.66 |
ENSRNOT00000026156
|
P2rx5
|
purinergic receptor P2X 5 |
| chr19_+_52664322 | 3.62 |
ENSRNOT00000082754
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr5_-_138239306 | 3.61 |
ENSRNOT00000039305
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_163570803 | 3.61 |
ENSRNOT00000082002
ENSRNOT00000078642 |
Klri2
|
killer cell lectin-like receptor family I member 2 |
| chr4_+_7662019 | 3.60 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr14_+_19319299 | 3.59 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_-_141307233 | 3.56 |
ENSRNOT00000071885
|
Racgap1
|
Rac GTPase-activating protein 1 |
| chr14_-_82347679 | 3.56 |
ENSRNOT00000032972
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr16_+_73584521 | 3.54 |
ENSRNOT00000024301
|
Gins4
|
GINS complex subunit 4 |
| chr4_-_163423628 | 3.54 |
ENSRNOT00000079526
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr2_-_165591110 | 3.54 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr6_-_138536321 | 3.53 |
ENSRNOT00000077743
|
AABR07065643.1
|
|
| chr12_-_20276121 | 3.51 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr4_+_93888502 | 3.49 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr3_+_91191837 | 3.49 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr8_+_103938520 | 3.49 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chrX_-_76924362 | 3.48 |
ENSRNOT00000078997
|
Atrx
|
ATRX, chromatin remodeler |
| chr5_-_161875362 | 3.48 |
ENSRNOT00000078237
|
Prdm2
|
PR/SET domain 2 |
| chr9_-_92405117 | 3.48 |
ENSRNOT00000079452
|
Trip12
|
thyroid hormone receptor interactor 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Olig2_Olig3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 3.5 | 10.6 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 3.2 | 9.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 3.1 | 18.4 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 3.0 | 8.9 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 2.9 | 8.7 | GO:1901580 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 2.8 | 8.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 2.7 | 10.7 | GO:1901423 | response to benzene(GO:1901423) |
| 2.5 | 10.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 2.4 | 9.6 | GO:0051542 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.4 | 9.5 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 2.0 | 9.8 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 1.9 | 11.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.8 | 9.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.6 | 9.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.6 | 6.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.6 | 4.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 1.6 | 10.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.5 | 4.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.4 | 4.2 | GO:0090579 | regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.4 | 5.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.3 | 4.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.3 | 8.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.3 | 13.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.2 | 3.7 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.2 | 7.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.2 | 3.6 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 1.2 | 4.8 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 1.2 | 3.6 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.2 | 4.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.1 | 9.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 3.3 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.1 | 3.2 | GO:1904760 | myofibroblast differentiation(GO:0036446) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) regulation of myofibroblast differentiation(GO:1904760) |
| 1.0 | 5.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.0 | 4.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.0 | 14.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.0 | 3.9 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.0 | 2.9 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 1.0 | 2.9 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.9 | 10.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.9 | 3.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.9 | 2.6 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.9 | 3.5 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.9 | 6.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.9 | 2.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.9 | 7.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.8 | 3.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.8 | 3.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.8 | 2.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 2.2 | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.7 | 2.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 3.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.7 | 3.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.7 | 2.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.6 | 3.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.6 | 11.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.6 | 2.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.6 | 2.5 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.6 | 2.4 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.6 | 1.8 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.6 | 1.2 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.6 | 13.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.6 | 10.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.5 | 2.7 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.5 | 2.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.5 | 2.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.5 | 10.0 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.5 | 5.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 1.5 | GO:0097213 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) positive regulation of protein folding(GO:1903334) |
| 0.5 | 4.7 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.5 | 2.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 1.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.4 | 6.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 4.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.4 | 3.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.4 | 1.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 1.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 3.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 2.8 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.4 | 2.8 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 2.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 1.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 3.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 1.2 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.4 | 2.7 | GO:0070782 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 7.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.4 | 2.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 5.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 5.6 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.4 | 1.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.4 | 2.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.4 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.3 | 9.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 4.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.3 | 3.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 5.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.3 | 3.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 1.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 11.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 5.4 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.3 | 3.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 1.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 0.9 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.3 | 1.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 3.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.3 | 8.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 1.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 2.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 4.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 3.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 4.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 2.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.8 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.3 | 0.8 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.3 | 1.6 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.3 | 2.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.3 | 1.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.3 | 0.5 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.2 | 10.7 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.2 | 4.7 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 2.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 11.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 2.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 8.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 1.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 3.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 14.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 0.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.9 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.2 | 4.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 2.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 5.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 3.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 5.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 4.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 1.0 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.2 | 0.6 | GO:0030578 | PML body organization(GO:0030578) cellular response to anoxia(GO:0071454) |
| 0.2 | 2.2 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 2.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 8.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 1.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 1.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 4.5 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 2.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) paranodal junction assembly(GO:0030913) |
| 0.2 | 1.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.7 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 0.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 1.4 | GO:0035635 | exocyst assembly(GO:0001927) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 5.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.2 | 4.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 2.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 5.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 3.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 | 3.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 1.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 6.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 8.0 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 3.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 9.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.0 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 1.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 4.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 1.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 3.9 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.6 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 1.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 5.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.6 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 1.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 4.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 0.7 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 3.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 10.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.6 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 1.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 4.9 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 0.3 | GO:0000393 | generation of catalytic spliceosome for second transesterification step(GO:0000350) spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 5.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 3.3 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.1 | 0.4 | GO:0051306 | mitotic sister chromatid separation(GO:0051306) |
| 0.1 | 2.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 1.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.6 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.3 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.6 | GO:0042045 | S-adenosylmethionine cycle(GO:0033353) epithelial fluid transport(GO:0042045) |
| 0.1 | 3.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.5 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.1 | 1.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 4.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 9.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.9 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 2.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 1.3 | GO:0051983 | regulation of chromosome segregation(GO:0051983) |
| 0.1 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 0.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 3.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.8 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.1 | 2.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 1.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 2.2 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 3.6 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.9 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 1.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.0 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 1.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 1.8 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.9 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.4 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 3.0 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.2 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 2.2 | 8.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.9 | 13.2 | GO:0042825 | TAP complex(GO:0042825) |
| 1.3 | 3.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.3 | 3.9 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.2 | 10.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.2 | 3.5 | GO:0000811 | GINS complex(GO:0000811) |
| 1.1 | 9.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.1 | 3.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 1.1 | 5.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.1 | 6.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.0 | 8.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.0 | 7.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 7.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.9 | 3.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.8 | 4.2 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.8 | 12.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.8 | 6.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 2.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.7 | 4.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.7 | 5.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.6 | 9.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 1.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.5 | 4.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.5 | 5.6 | GO:0002177 | manchette(GO:0002177) |
| 0.5 | 1.5 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.5 | 3.0 | GO:0036157 | outer dynein arm(GO:0036157) sperm cytoplasmic droplet(GO:0097598) |
| 0.5 | 2.9 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.5 | 1.9 | GO:0097196 | Shu complex(GO:0097196) |
| 0.4 | 11.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 10.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 2.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 9.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 8.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 7.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 2.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 9.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 2.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 1.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 3.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 0.9 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 1.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 3.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 3.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 4.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 16.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 4.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 2.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 1.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 11.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 6.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 44.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 21.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 3.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 5.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 6.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 14.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 5.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 5.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 8.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 28.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 4.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 7.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 10.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 14.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 6.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 2.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 3.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 5.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0032153 | actomyosin contractile ring(GO:0005826) cell division site(GO:0032153) cell division site part(GO:0032155) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 4.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 2.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 2.7 | 8.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.6 | 13.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 1.9 | 9.6 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.9 | 7.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.7 | 8.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.7 | 5.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.4 | 9.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.3 | 4.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.3 | 3.9 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.2 | 3.6 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 1.2 | 10.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.2 | 4.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.1 | 3.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.1 | 8.9 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 1.1 | 3.3 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.1 | 3.2 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 1.0 | 6.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.0 | 3.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 1.0 | 11.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.0 | 2.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.9 | 1.8 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.9 | 9.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.9 | 5.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 6.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.7 | 2.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.7 | 2.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.7 | 14.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.6 | 3.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.6 | 4.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 1.3 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.6 | 1.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 4.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.6 | 5.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.6 | 7.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 1.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 3.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.5 | 5.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 1.5 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.5 | 2.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.5 | 4.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 8.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 2.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 13.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.5 | 4.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 2.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 5.8 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 2.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.4 | 4.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 2.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 9.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.4 | 4.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 3.7 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 8.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.4 | 2.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 1.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 2.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.4 | 5.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.8 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.3 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 12.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 2.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.3 | 9.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 1.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 1.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.3 | 3.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 3.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 7.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 15.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 2.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 3.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 6.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 3.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 5.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 2.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 2.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 2.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 32.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 5.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 3.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 0.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.1 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.1 | 2.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 11.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.5 | GO:0004383 | adenylate cyclase activity(GO:0004016) guanylate cyclase activity(GO:0004383) |
| 0.1 | 5.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 3.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 8.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 5.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 32.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 22.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 5.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 27.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 2.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 11.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 6.7 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 3.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 31.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 3.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.5 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.8 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 3.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 3.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 3.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 7.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 12.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 7.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 2.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 1.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 17.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0052813 | insulin receptor substrate binding(GO:0043560) phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.5 | 2.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 4.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 10.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 13.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 10.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 3.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 14.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 3.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 7.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 2.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 5.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 16.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 6.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 2.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 8.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 6.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 9.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 14.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 8.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 3.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 8.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 5.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.6 | 5.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 10.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 21.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.5 | 9.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.5 | 11.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 13.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 5.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 22.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 9.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.4 | 4.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 9.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 4.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 2.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 9.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 3.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 14.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 4.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 12.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 3.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 5.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 5.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 8.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 9.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 10.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 4.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 8.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 8.0 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 10.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 3.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 5.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 0.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.0 | 5.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |