Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
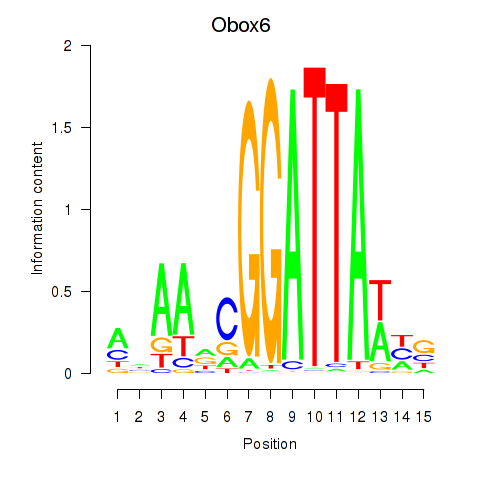
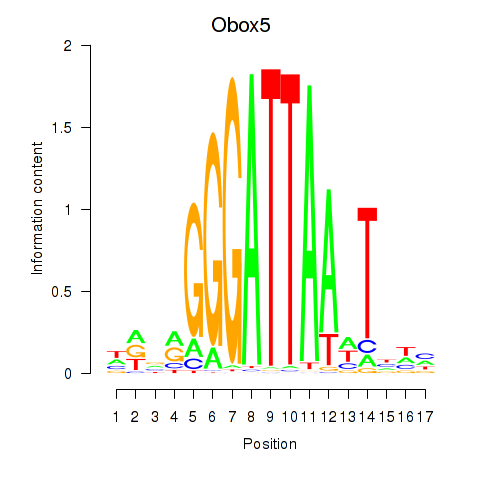
Results for Obox6_Obox5
Z-value: 0.96
Transcription factors associated with Obox6_Obox5
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Obox6_Obox5 motif
Sorted Z-values of Obox6_Obox5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_25749098 | 48.24 |
ENSRNOT00000027771
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr20_+_28572242 | 34.46 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr11_-_4397361 | 32.99 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr1_-_67284864 | 25.72 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr3_+_113318563 | 21.92 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_-_106753592 | 21.82 |
ENSRNOT00000006930
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr11_+_57505005 | 21.82 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr16_+_39144972 | 20.02 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr3_+_48106099 | 19.99 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr8_-_53899669 | 19.94 |
ENSRNOT00000082257
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr11_-_59110562 | 19.54 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr7_-_124929025 | 19.02 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr14_-_115052450 | 18.28 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr6_-_23291568 | 17.52 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr14_+_8182383 | 17.40 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr16_+_39145230 | 17.36 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr14_+_81858737 | 17.32 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr1_+_67025240 | 16.63 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr11_+_57207656 | 16.56 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_+_5930298 | 16.47 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr2_-_115836846 | 16.42 |
ENSRNOT00000014359
|
Cldn11
|
claudin 11 |
| chr16_+_39353283 | 16.15 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr11_+_20471764 | 15.99 |
ENSRNOT00000078127
|
Ncam2
|
neural cell adhesion molecule 2 |
| chr7_+_133457255 | 15.22 |
ENSRNOT00000080621
|
Cntn1
|
contactin 1 |
| chr14_+_39663421 | 15.20 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr17_+_81455955 | 14.69 |
ENSRNOT00000044313
|
Slc39a12
|
solute carrier family 39 member 12 |
| chr3_+_56862691 | 14.63 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr7_+_27620458 | 14.58 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr11_+_20474483 | 14.41 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr12_-_10335499 | 14.25 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chrX_-_51792597 | 13.96 |
ENSRNOT00000072727
|
LOC102549011
|
titin-like |
| chr10_-_27862868 | 13.71 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr18_-_67224566 | 13.69 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr15_+_17775692 | 13.69 |
ENSRNOT00000061169
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr4_+_84423653 | 13.69 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr18_-_74485139 | 13.68 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr6_+_106084815 | 13.66 |
ENSRNOT00000058195
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_4332255 | 13.32 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr2_+_92573004 | 13.30 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr17_+_23661429 | 13.25 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr6_-_14523961 | 13.20 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr19_-_57192095 | 13.16 |
ENSRNOT00000058080
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr1_-_211196868 | 13.05 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr1_+_212714179 | 12.75 |
ENSRNOT00000054880
ENSRNOT00000025680 |
Olr292
|
olfactory receptor 292 |
| chr15_-_29623597 | 12.64 |
ENSRNOT00000074483
|
AABR07017647.1
|
|
| chr20_-_8574082 | 12.46 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr1_+_100393303 | 12.39 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr13_+_6679368 | 12.38 |
ENSRNOT00000067198
ENSRNOT00000092965 |
Cntnap5c
|
contactin associated protein-like 5C |
| chr6_+_27328406 | 12.30 |
ENSRNOT00000091159
ENSRNOT00000044278 |
Otof
|
otoferlin |
| chr1_-_54854353 | 12.19 |
ENSRNOT00000072895
|
Smok2a
|
sperm motility kinase 2A |
| chr15_+_47470863 | 12.11 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr19_+_20607507 | 12.02 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr20_+_27592379 | 12.02 |
ENSRNOT00000047000
|
Trappc3l
|
trafficking protein particle complex 3-like |
| chr18_-_86878142 | 11.97 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chr14_-_39112600 | 11.85 |
ENSRNOT00000003170
|
Gabrb1
|
gamma-aminobutyric acid type A receptor beta 1 subunit |
| chr6_-_54059119 | 11.83 |
ENSRNOT00000005521
|
Hdac9
|
histone deacetylase 9 |
| chr16_-_54628458 | 11.70 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr1_-_67206713 | 11.63 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chr13_+_50103189 | 11.63 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr1_-_261333383 | 11.61 |
ENSRNOT00000037490
|
Morn4
|
MORN repeat containing 4 |
| chr9_-_30844199 | 11.34 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr18_+_54108493 | 11.32 |
ENSRNOT00000029239
|
RGD1312005
|
similar to DD1 |
| chrX_-_15620841 | 11.26 |
ENSRNOT00000085397
|
Praf2
|
PRA1 domain family, member 2 |
| chr4_-_128266082 | 11.23 |
ENSRNOT00000039549
|
Nup50
|
nucleoporin 50 |
| chr10_+_92288910 | 11.22 |
ENSRNOT00000006947
ENSRNOT00000045127 |
Mapt
|
microtubule-associated protein tau |
| chrX_-_157286936 | 11.21 |
ENSRNOT00000078100
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr15_+_17834635 | 11.11 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr11_+_66316606 | 11.07 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr7_+_38897278 | 11.06 |
ENSRNOT00000006450
|
Epyc
|
epiphycan |
| chr7_+_20262680 | 11.05 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chrX_+_122808605 | 11.02 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr17_-_61332391 | 11.02 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr7_-_117732339 | 10.91 |
ENSRNOT00000092917
ENSRNOT00000020696 |
Foxh1
|
forkhead box H1 |
| chr1_+_72661211 | 10.89 |
ENSRNOT00000033197
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr9_+_116652530 | 10.87 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr9_+_12114977 | 10.70 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr3_+_98297554 | 10.69 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr1_+_78893271 | 10.68 |
ENSRNOT00000029825
|
Pnmal1
|
paraneoplastic Ma antigen family-like 1 |
| chr14_-_113644779 | 10.60 |
ENSRNOT00000005306
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr4_-_70934295 | 10.48 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr1_+_192379543 | 10.47 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr3_-_12155098 | 10.35 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_+_143151739 | 10.20 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chr4_-_155763500 | 10.18 |
ENSRNOT00000071745
|
LOC100909595
|
solute carrier family 2, facilitated glucose transporter member 3-like |
| chr5_-_32956159 | 10.12 |
ENSRNOT00000078264
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr3_-_102151489 | 10.01 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr5_-_158549496 | 9.97 |
ENSRNOT00000074336
|
Igsf21
|
immunoglobin superfamily, member 21 |
| chr9_+_8349033 | 9.93 |
ENSRNOT00000073775
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr4_+_140837745 | 9.89 |
ENSRNOT00000080663
|
Arl8b
|
ADP-ribosylation factor like GTPase 8B |
| chr7_+_139685573 | 9.86 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chrX_+_20423401 | 9.78 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr1_+_87563975 | 9.71 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr6_-_104409005 | 9.70 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr13_+_77602249 | 9.67 |
ENSRNOT00000003407
ENSRNOT00000076589 |
Tnr
|
tenascin R |
| chr3_-_52664209 | 9.67 |
ENSRNOT00000065126
ENSRNOT00000079020 |
Scn9a
|
sodium voltage-gated channel alpha subunit 9 |
| chr4_+_9981958 | 9.63 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr17_-_65955606 | 9.52 |
ENSRNOT00000067949
ENSRNOT00000023601 |
Ryr2
|
ryanodine receptor 2 |
| chr1_-_266074181 | 9.46 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr6_+_100337226 | 9.34 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr9_+_73418607 | 9.33 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chrX_-_115764217 | 9.26 |
ENSRNOT00000009400
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr14_+_113867209 | 9.20 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr10_-_12916784 | 9.13 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr1_-_72335855 | 9.09 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr3_+_63379031 | 9.08 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr3_-_46153371 | 9.02 |
ENSRNOT00000085885
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_-_188561267 | 8.97 |
ENSRNOT00000089781
ENSRNOT00000092093 |
Trim46
|
tripartite motif-containing 46 |
| chr7_+_123482255 | 8.94 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr19_+_10731855 | 8.93 |
ENSRNOT00000022277
|
Pllp
|
plasmolipin |
| chr17_-_71105286 | 8.88 |
ENSRNOT00000025901
|
Prkcq
|
protein kinase C, theta |
| chr1_-_84950210 | 8.86 |
ENSRNOT00000083050
|
AABR07002784.1
|
|
| chr2_+_54191538 | 8.83 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_-_50923510 | 8.81 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr6_-_95061174 | 8.78 |
ENSRNOT00000072262
|
Rtn1
|
reticulon 1 |
| chr2_+_115337439 | 8.76 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr6_-_127319362 | 8.76 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr4_+_153330069 | 8.71 |
ENSRNOT00000015560
|
Slc25a18
|
solute carrier family 25 member 18 |
| chr11_-_88180542 | 8.69 |
ENSRNOT00000002539
|
Ypel1
|
yippee-like 1 |
| chr4_+_91373942 | 8.62 |
ENSRNOT00000043827
|
Ccser1
|
coiled-coil serine-rich protein 1 |
| chrY_+_914045 | 8.58 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr13_-_91872954 | 8.45 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr7_+_129595192 | 8.45 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr19_+_14835822 | 8.43 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr10_-_49192693 | 8.30 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chr8_+_61080103 | 8.26 |
ENSRNOT00000022976
|
Hmg20a
|
high mobility group 20A |
| chr5_+_159612762 | 8.23 |
ENSRNOT00000012147
|
Spata21
|
spermatogenesis associated 21 |
| chr4_-_51586881 | 8.21 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr7_+_92234597 | 8.14 |
ENSRNOT00000067491
ENSRNOT00000089991 |
Med30
|
mediator complex subunit 30 |
| chr13_+_71107465 | 8.11 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr7_+_15785410 | 8.08 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr15_+_31417147 | 8.04 |
ENSRNOT00000092182
|
AABR07017824.2
|
|
| chr4_-_177196180 | 8.01 |
ENSRNOT00000018999
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr14_+_115275894 | 8.00 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr1_-_4011161 | 7.97 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr6_+_10483308 | 7.89 |
ENSRNOT00000074516
|
Tmem247
|
transmembrane protein 247 |
| chr10_+_69737328 | 7.88 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr18_-_37986930 | 7.80 |
ENSRNOT00000025927
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr6_-_8344897 | 7.78 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr3_+_170994038 | 7.78 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr2_+_66940057 | 7.67 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr5_+_122390522 | 7.67 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr14_-_3288017 | 7.67 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr2_-_186606172 | 7.60 |
ENSRNOT00000021938
|
Fcrl2
|
Fc receptor-like 2 |
| chr20_+_13498926 | 7.56 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr8_-_84506328 | 7.56 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr20_+_41100071 | 7.52 |
ENSRNOT00000000658
|
Tspyl4
|
TSPY-like 4 |
| chrX_-_37003642 | 7.39 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr12_-_10391270 | 7.38 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr19_-_25378974 | 7.38 |
ENSRNOT00000011150
|
Ccdc130
|
coiled-coil domain containing 130 |
| chr10_+_93305969 | 7.34 |
ENSRNOT00000008019
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr1_-_67302751 | 7.32 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr11_+_74984613 | 7.24 |
ENSRNOT00000035049
|
Atp13a5
|
ATPase 13A5 |
| chr1_-_174119815 | 7.20 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr14_+_22091777 | 7.19 |
ENSRNOT00000072483
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr1_+_234252757 | 7.15 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr15_-_71779033 | 7.13 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr5_-_65073012 | 7.13 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr6_-_114476723 | 7.09 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr8_+_23398030 | 7.04 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr15_+_47455690 | 6.96 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr16_+_52024690 | 6.94 |
ENSRNOT00000048104
|
AABR07025825.1
|
|
| chr8_+_64481172 | 6.91 |
ENSRNOT00000015332
|
Pkm
|
pyruvate kinase, muscle |
| chr18_-_27395603 | 6.83 |
ENSRNOT00000034385
|
LOC100359679
|
rCG49431-like |
| chr1_+_266781617 | 6.81 |
ENSRNOT00000027417
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr14_+_17534412 | 6.78 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr3_-_15278645 | 6.76 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr7_-_107203897 | 6.72 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr2_+_144646308 | 6.68 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_-_176983045 | 6.66 |
ENSRNOT00000022301
|
Dkk3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_-_4550768 | 6.65 |
ENSRNOT00000061954
|
Adgb
|
androglobin |
| chr7_-_139872900 | 6.60 |
ENSRNOT00000046402
|
H1fnt
|
H1 histone family, member N, testis-specific |
| chrX_-_43781586 | 6.58 |
ENSRNOT00000051551
|
Cldn34b
|
claudin 34B |
| chr1_+_48077033 | 6.53 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr14_-_80130139 | 6.45 |
ENSRNOT00000091652
ENSRNOT00000010482 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr15_-_29761117 | 6.43 |
ENSRNOT00000075194
|
AABR07017658.2
|
|
| chr3_+_56030915 | 6.41 |
ENSRNOT00000010489
|
Phospho2
|
phosphatase, orphan 2 |
| chrX_-_60852583 | 6.40 |
ENSRNOT00000004145
|
Mageb5
|
MAGE family member B5 |
| chr1_-_100359511 | 6.39 |
ENSRNOT00000056420
|
RGD1309036
|
hypothetical LOC292874 |
| chr13_-_113817995 | 6.33 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chrX_+_83570346 | 6.30 |
ENSRNOT00000045603
|
Ube2d4l1
|
ubiquitin-conjugating enzyme E2D 4 like 1 |
| chr5_+_59178846 | 6.28 |
ENSRNOT00000021227
|
Tmem8b
|
transmembrane protein 8B |
| chr18_+_30496318 | 6.25 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr2_+_18354542 | 6.24 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr8_-_13513337 | 6.23 |
ENSRNOT00000071532
|
LOC108348070
|
lysine-specific demethylase 4D |
| chr4_-_129430251 | 6.20 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr19_+_37668133 | 6.11 |
ENSRNOT00000044971
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr1_-_72464492 | 6.10 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr9_+_73334618 | 6.09 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr17_+_35396286 | 6.04 |
ENSRNOT00000089613
ENSRNOT00000092638 |
Exoc2
|
exocyst complex component 2 |
| chr18_+_57654290 | 6.00 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr2_+_207262934 | 6.00 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr1_+_71252950 | 6.00 |
ENSRNOT00000067530
|
Zfp583
|
zinc finger protein 583 |
| chr5_+_8876044 | 5.97 |
ENSRNOT00000008877
|
Cops5
|
COP9 signalosome subunit 5 |
| chr7_-_116106368 | 5.91 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr8_+_129205931 | 5.89 |
ENSRNOT00000082377
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr5_+_64326733 | 5.88 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr5_+_146656049 | 5.88 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr3_+_143172686 | 5.86 |
ENSRNOT00000006869
|
Cst9l
|
cystatin 9-like |
| chr8_+_119083900 | 5.85 |
ENSRNOT00000051947
ENSRNOT00000048655 |
Prss44
|
protease, serine, 44 |
| chr11_-_71743421 | 5.84 |
ENSRNOT00000002395
|
Rnf168
|
ring finger protein 168 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox6_Obox5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 6.6 | 13.2 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 5.5 | 16.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 3.9 | 11.6 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 3.7 | 48.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 3.6 | 10.9 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 3.5 | 10.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.4 | 13.7 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 3.3 | 13.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.3 | 9.8 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 3.2 | 9.5 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 3.1 | 9.3 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 3.1 | 9.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 3.0 | 18.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 3.0 | 8.9 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 2.9 | 11.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 2.9 | 14.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.8 | 11.1 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 2.6 | 10.3 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 2.5 | 53.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 2.5 | 12.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 2.5 | 9.9 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 2.4 | 9.6 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 2.4 | 11.9 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 2.3 | 18.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 2.3 | 9.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.1 | 12.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.9 | 3.9 | GO:0002856 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 1.9 | 5.8 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 1.9 | 15.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.9 | 5.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.8 | 5.5 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 1.8 | 9.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 1.7 | 18.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.7 | 10.0 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.7 | 6.7 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 1.6 | 8.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 1.5 | 4.6 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.5 | 10.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 1.5 | 16.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.5 | 8.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.5 | 8.8 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 1.4 | 5.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.4 | 11.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.4 | 4.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 1.4 | 8.1 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 1.3 | 14.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.3 | 5.3 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 1.3 | 4.0 | GO:1901660 | calcium ion export(GO:1901660) |
| 1.2 | 2.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 1.2 | 6.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 1.2 | 4.9 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 1.2 | 12.0 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.2 | 4.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.2 | 11.8 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.2 | 49.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.2 | 3.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.2 | 12.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 1.1 | 2.3 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 1.1 | 6.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.1 | 17.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 1.1 | 12.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.0 | 7.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.0 | 30.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 1.0 | 52.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 1.0 | 1.0 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 1.0 | 3.9 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 1.0 | 16.7 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 1.0 | 18.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 1.0 | 2.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.9 | 6.6 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.9 | 3.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.9 | 8.4 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.9 | 2.8 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.9 | 3.7 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.9 | 3.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.9 | 1.8 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.9 | 9.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.9 | 5.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.8 | 5.8 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.8 | 13.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.8 | 2.4 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.8 | 7.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.8 | 2.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 6.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.8 | 4.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.7 | 2.9 | GO:0050955 | thermoception(GO:0050955) |
| 0.7 | 2.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 8.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.7 | 2.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.7 | 2.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.7 | 12.8 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.7 | 7.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.7 | 7.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.7 | 6.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.7 | 6.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.7 | 4.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 2.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.7 | 2.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 4.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.6 | 12.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.6 | 12.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.6 | 2.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.6 | 1.8 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.6 | 4.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.6 | 2.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.6 | 2.9 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.6 | 3.4 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.6 | 5.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.6 | 5.6 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.6 | 3.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.6 | 0.6 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.5 | 6.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.5 | 17.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.5 | 1.5 | GO:0015676 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 0.5 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 4.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 5.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.5 | 3.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 5.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 6.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 2.7 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 1.4 | GO:2000424 | negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.4 | 2.7 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 3.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.4 | 3.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 15.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 6.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.4 | 5.9 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.4 | 5.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.4 | 1.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.4 | 3.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.4 | 2.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 1.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.4 | 1.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.4 | 1.9 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.4 | 4.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.4 | 1.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.4 | 1.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.4 | 1.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.4 | 1.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 8.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 3.2 | GO:0044791 | negative regulation of DNA endoreduplication(GO:0032876) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.4 | 1.8 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) mesonephric duct formation(GO:0072181) |
| 0.4 | 3.9 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.4 | 13.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 1.4 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.3 | 1.0 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.3 | 1.7 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.3 | 6.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 7.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.3 | 2.0 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 7.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.3 | 1.0 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 | 4.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.6 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.3 | 15.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.3 | 1.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.3 | 1.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.3 | 3.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 2.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 14.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.3 | 2.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.3 | 1.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.3 | 0.8 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.3 | 3.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.3 | 7.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 | 2.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.9 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.3 | 2.6 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) |
| 0.2 | 19.4 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.2 | 6.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 5.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 4.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 1.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.2 | 1.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 3.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 7.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 1.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 0.5 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.2 | 1.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 0.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 0.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 2.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.6 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.0 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.2 | 3.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.2 | 7.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 16.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.2 | 0.6 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 4.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 0.7 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.2 | 15.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.2 | 3.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 3.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.2 | 8.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.2 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.2 | 19.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 5.9 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.2 | 3.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 7.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.2 | 0.7 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.2 | 0.5 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 2.7 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 1.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 1.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 2.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.9 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) neutrophil differentiation(GO:0030223) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.9 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 2.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 1.6 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 1.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 0.3 | GO:0052564 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) response to host(GO:0075136) |
| 0.1 | 0.9 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.9 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 0.8 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.8 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 10.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 1.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 2.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 8.6 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 1.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.3 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.1 | 4.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 3.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 3.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.8 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 2.5 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 2.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 1.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 2.5 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 2.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 0.9 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 2.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 4.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.8 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 2.4 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.9 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 2.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.2 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 1.7 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 3.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.8 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 1.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 4.9 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 1.8 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0070340 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 2.4 | GO:0048588 | developmental cell growth(GO:0048588) |
| 0.0 | 4.0 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 3.6 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.0 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 2.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 2.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 35.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 3.3 | 9.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 2.8 | 11.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.9 | 9.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.8 | 10.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 1.8 | 9.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.7 | 13.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.6 | 37.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.6 | 27.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.6 | 6.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 1.5 | 5.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.4 | 54.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 1.4 | 5.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.2 | 12.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.2 | 5.9 | GO:0098536 | deuterosome(GO:0098536) |
| 1.1 | 27.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.1 | 8.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.0 | 12.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 1.0 | 3.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.9 | 5.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 2.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.8 | 16.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.8 | 9.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 12.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.7 | 2.7 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.6 | 1.9 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.6 | 10.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.6 | 1.8 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.6 | 27.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.6 | 10.0 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.6 | 3.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.6 | 1.7 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.6 | 2.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.6 | 4.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.6 | 20.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.6 | 2.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 33.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.6 | 6.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.6 | 8.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.5 | 2.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.5 | 1.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.5 | 6.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.5 | 3.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 1.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.5 | 2.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.5 | 5.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 1.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 1.9 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.5 | 3.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 9.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 6.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 3.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 2.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 13.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 3.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 1.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.4 | 4.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.4 | 15.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 4.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 2.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.4 | 5.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 2.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.4 | 8.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.4 | 11.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 5.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 4.1 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 7.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 3.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 2.1 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 8.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 19.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 6.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 51.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 9.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 9.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 2.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 1.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 5.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 1.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.3 | 1.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 2.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 1.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.3 | 3.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 45.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.3 | 33.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 0.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.3 | 5.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 7.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 11.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 2.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 7.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 4.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 2.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 10.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 1.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 27.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 33.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 12.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 5.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 8.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 1.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 4.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.1 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.2 | 0.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 17.0 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 72.1 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 7.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 19.0 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 1.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 12.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 3.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 4.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 6.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 15.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 3.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 5.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.2 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 5.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 19.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 4.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.9 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 12.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 4.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 3.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 13.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 3.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 28.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 5.2 | 10.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 4.6 | 13.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 4.5 | 18.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 4.1 | 49.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 3.1 | 21.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.0 | 11.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 2.9 | 14.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.8 | 11.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.7 | 13.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 2.5 | 9.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 2.5 | 12.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.4 | 9.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.4 | 23.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 2.0 | 6.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 1.9 | 7.8 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.9 | 13.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.9 | 18.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.7 | 8.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.7 | 6.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.7 | 3.4 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 1.6 | 9.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.5 | 15.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 1.5 | 10.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.4 | 5.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.4 | 4.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 1.4 | 4.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.3 | 21.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 1.3 | 5.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.2 | 15.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 1.2 | 14.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.1 | 12.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 1.1 | 4.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.0 | 7.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.0 | 4.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.0 | 5.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.9 | 6.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.9 | 18.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.9 | 15.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.9 | 4.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.9 | 2.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.8 | 19.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.8 | 12.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 3.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.8 | 9.5 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.8 | 3.9 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.8 | 2.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.8 | 5.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 9.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.8 | 7.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 10.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.7 | 2.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.7 | 2.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.7 | 7.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.7 | 4.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.7 | 7.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.7 | 3.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 9.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.6 | 7.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.6 | 3.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.6 | 12.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 10.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.6 | 10.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.6 | 2.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.6 | 12.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.6 | 4.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 2.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.6 | 3.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.6 | 56.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.6 | 1.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 16.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.6 | 12.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 15.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.5 | 9.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.5 | 66.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 15.3 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.5 | 1.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.5 | 1.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 7.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.5 | 1.8 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.5 | 13.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 4.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 1.8 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.5 | 9.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 1.8 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.4 | 4.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 14.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 1.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 1.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.4 | 1.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.4 | 8.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 13.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 1.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 4.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 16.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.4 | 11.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 2.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.4 | 3.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 2.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.3 | 8.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.3 | 1.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.3 | 1.0 | GO:0035596 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.3 | 12.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 2.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 6.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 13.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 2.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 8.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 7.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 8.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 0.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 1.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 1.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.3 | 3.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 12.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 10.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.3 | 2.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 4.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 5.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 25.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.3 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 8.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 6.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 21.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 2.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 3.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 3.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 8.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 28.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 5.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.7 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.2 | 6.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.6 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.2 | 4.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 4.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 2.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 0.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 3.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 1.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 0.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 3.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.2 | 4.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.8 | GO:0090079 | translation repressor activity, nucleic acid binding(GO:0000900) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.1 | 2.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 34.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.0 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.7 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 8.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 6.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 6.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 8.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 14.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 8.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 4.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 29.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 2.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.3 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 2.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.6 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 2.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 1.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 7.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0042498 | Toll-like receptor 2 binding(GO:0035663) diacyl lipopeptide binding(GO:0042498) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 62.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.9 | 11.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 10.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 27.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.6 | 16.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.5 | 27.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.5 | 13.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 17.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 9.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.4 | 19.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 5.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 14.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 15.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 15.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 6.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 5.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 3.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 5.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 8.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 6.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 5.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 7.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 12.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 2.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 23.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 40.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.9 | 52.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.9 | 71.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 1.8 | 23.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 1.2 | 18.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.8 | 10.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.8 | 15.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.8 | 10.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.7 | 19.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.6 | 22.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 22.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.6 | 8.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 14.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.5 | 12.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.5 | 18.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.5 | 10.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 14.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.4 | 9.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.4 | 4.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 8.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 9.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 4.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 10.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 7.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 2.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 4.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 4.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 5.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 15.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 9.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 11.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.8 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.2 | 10.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 7.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 5.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 4.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 8.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 14.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 3.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 5.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 17.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 6.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 3.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 6.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 2.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |