Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
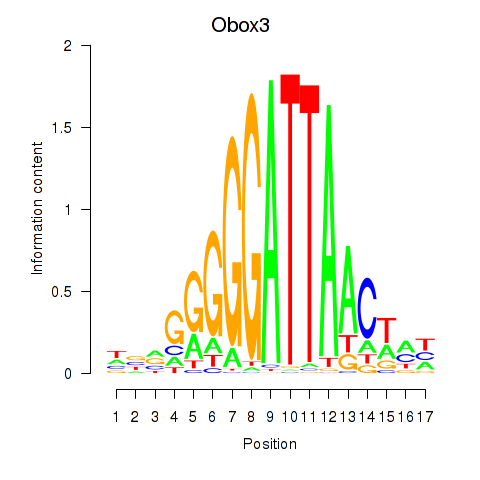
Results for Obox3
Z-value: 0.51
Transcription factors associated with Obox3
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RGD1564236 | rn6_v1_chr9_+_101197255_101197255 | 0.44 | 2.6e-16 | Click! |
Activity profile of Obox3 motif
Sorted Z-values of Obox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_51576376 | 16.18 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr20_-_8574082 | 12.10 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr18_-_37776453 | 11.26 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr10_-_55560422 | 11.16 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr4_+_123586231 | 11.07 |
ENSRNOT00000049657
ENSRNOT00000013232 |
Grip2
|
glutamate receptor interacting protein 2 |
| chr10_+_83954279 | 10.38 |
ENSRNOT00000006594
|
Ttll6
|
tubulin tyrosine ligase like 6 |
| chr6_-_8344897 | 10.24 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr6_+_10483308 | 9.17 |
ENSRNOT00000074516
|
Tmem247
|
transmembrane protein 247 |
| chr3_+_43255567 | 8.94 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr4_-_155763500 | 8.58 |
ENSRNOT00000071745
|
LOC100909595
|
solute carrier family 2, facilitated glucose transporter member 3-like |
| chr3_+_112242270 | 8.24 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr8_-_87315955 | 7.90 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr17_-_57526461 | 7.25 |
ENSRNOT00000072723
|
LOC103690053
|
ankyrin repeat domain-containing protein 26-like |
| chr7_-_49741540 | 6.88 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr2_-_62634785 | 6.56 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr2_+_93712992 | 6.53 |
ENSRNOT00000059326
|
Fabp12
|
fatty acid binding protein 12 |
| chr7_+_94795214 | 6.52 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr19_+_51985170 | 6.37 |
ENSRNOT00000019443
|
Hsbp1
|
heat shock factor binding protein 1 |
| chr1_+_224824799 | 6.14 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chrX_-_73562046 | 6.13 |
ENSRNOT00000080501
|
Gm14597
|
predicted gene 14597 |
| chr1_+_72661211 | 6.12 |
ENSRNOT00000033197
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr14_+_23089699 | 5.79 |
ENSRNOT00000036948
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr6_-_106971250 | 5.39 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr12_-_22477052 | 5.36 |
ENSRNOT00000075504
|
Ache
|
acetylcholinesterase |
| chr8_-_107490093 | 5.01 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr2_-_41785792 | 4.94 |
ENSRNOT00000015871
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chrM_+_7758 | 4.58 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chrM_+_7919 | 4.57 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_+_133896141 | 4.54 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_-_26855658 | 4.51 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr6_+_101288951 | 4.36 |
ENSRNOT00000046901
|
RGD1562540
|
RGD1562540 |
| chrX_-_116638508 | 4.34 |
ENSRNOT00000030400
|
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr10_+_15156207 | 3.98 |
ENSRNOT00000020363
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr10_+_69737328 | 3.90 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr4_-_100783750 | 3.78 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr10_-_83332851 | 3.64 |
ENSRNOT00000007133
|
Nxph3
|
neurexophilin 3 |
| chrX_+_105911925 | 3.43 |
ENSRNOT00000052422
|
LOC108348137
|
armadillo repeat-containing X-linked protein 1 |
| chr15_+_3938075 | 2.99 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr8_+_94686938 | 2.93 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr4_-_165747201 | 2.69 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr7_-_106695570 | 2.64 |
ENSRNOT00000083517
|
Hhla1
|
HERV-H LTR-associating 1 |
| chr10_+_54512983 | 2.43 |
ENSRNOT00000005204
|
Stx8
|
syntaxin 8 |
| chr5_+_81431600 | 2.29 |
ENSRNOT00000013675
|
Trim32
|
tripartite motif-containing 32 |
| chr13_-_35858390 | 2.27 |
ENSRNOT00000003568
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr5_+_159484370 | 2.25 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr19_+_60017746 | 2.17 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr13_+_77940454 | 2.17 |
ENSRNOT00000003460
|
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr16_+_19896986 | 2.00 |
ENSRNOT00000060355
|
Gtpbp3
|
GTP binding protein 3 |
| chr6_-_132958546 | 1.95 |
ENSRNOT00000041903
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr1_-_266074181 | 1.93 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr10_-_15305549 | 1.79 |
ENSRNOT00000027353
|
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr17_+_81798756 | 1.70 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_-_91807937 | 1.63 |
ENSRNOT00000005074
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr5_-_61734011 | 1.56 |
ENSRNOT00000012915
|
Tstd2
|
thiosulfate sulfurtransferase like domain containing 2 |
| chr8_-_116465801 | 1.43 |
ENSRNOT00000085915
|
Sema3f
|
semaphorin 3F |
| chr19_-_43215077 | 1.39 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr1_+_189870622 | 1.37 |
ENSRNOT00000075003
|
Tmem159
|
transmembrane protein 159 |
| chr5_+_98387291 | 1.23 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr18_+_40962146 | 1.16 |
ENSRNOT00000035656
|
Arl14epl
|
ADP ribosylation factor like GTPase 14 effector protein like |
| chr8_+_55050284 | 1.14 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr19_-_43215281 | 1.14 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr10_-_55851235 | 1.10 |
ENSRNOT00000010790
|
Gucy2d
|
guanylate cyclase 2D, retinal |
| chr3_+_75437833 | 1.07 |
ENSRNOT00000042246
|
Olr561
|
olfactory receptor 561 |
| chr1_+_213686046 | 1.03 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr15_+_41448064 | 1.03 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr4_-_153593773 | 0.69 |
ENSRNOT00000082982
|
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr5_-_115387377 | 0.69 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_-_22404002 | 0.55 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr19_+_9622611 | 0.28 |
ENSRNOT00000061498
|
Slc38a7
|
solute carrier family 38, member 7 |
| chr17_+_34665872 | 0.23 |
ENSRNOT00000092101
ENSRNOT00000085704 |
Exoc2
|
exocyst complex component 2 |
| chr10_-_103481153 | 0.20 |
ENSRNOT00000035639
|
Cd300lb
|
CD300 molecule-like family member b |
| chr18_+_56544652 | 0.12 |
ENSRNOT00000024171
|
Pde6a
|
phosphodiesterase 6A |
| chr8_+_41285450 | 0.06 |
ENSRNOT00000076562
|
Olr1222
|
olfactory receptor 1222 |
| chr2_+_60966789 | 0.05 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.7 | 8.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 2.0 | 12.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.8 | 8.9 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 1.2 | 11.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 1.0 | 10.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.0 | 6.1 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.9 | 6.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.9 | 8.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.8 | 11.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.8 | 11.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 2.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.6 | 5.4 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.6 | 4.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.6 | 1.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 6.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.5 | 4.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.4 | 2.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 2.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.6 | GO:0072181 | mesonephric duct formation(GO:0072181) |
| 0.3 | 1.9 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.3 | 9.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 1.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 3.0 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 1.4 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.2 | 2.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 6.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 2.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 2.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 6.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 4.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.8 | 4.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 2.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.6 | 9.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.4 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 6.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 5.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.4 | 5.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 12.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 16.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 11.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 17.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 8.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 8.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 10.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 4.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 8.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 7.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 13.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 14.5 | GO:0005856 | cytoskeleton(GO:0005856) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 2.8 | 11.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.1 | 10.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 1.8 | 5.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.4 | 11.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.0 | 8.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 10.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.8 | 2.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.6 | 2.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.6 | 6.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 8.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 11.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.4 | 2.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 1.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 13.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 6.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 4.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 2.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 4.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.9 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 1.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 9.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 3.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 2.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 6.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 3.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 6.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.6 | 11.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.6 | 6.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 6.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 13.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 8.9 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |