Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
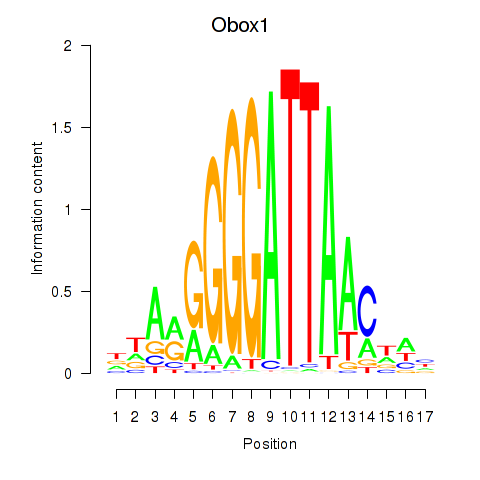
Results for Obox1
Z-value: 0.99
Transcription factors associated with Obox1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Obox1 motif
Sorted Z-values of Obox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_85645718 | 40.30 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chr1_-_189199376 | 39.70 |
ENSRNOT00000021027
|
Umod
|
uromodulin |
| chr2_+_54466280 | 34.13 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr2_+_199796881 | 32.17 |
ENSRNOT00000024423
|
Fmo5
|
flavin containing monooxygenase 5 |
| chr3_+_159936856 | 29.57 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr10_-_62273119 | 27.30 |
ENSRNOT00000004322
|
Serpinf2
|
serpin family F member 2 |
| chr14_+_22375955 | 27.28 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr4_-_30276372 | 24.35 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr11_+_82945104 | 24.01 |
ENSRNOT00000002410
|
Ehhadh
|
enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase |
| chr7_+_141370491 | 23.60 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr15_-_28104206 | 23.12 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr3_-_48372583 | 22.69 |
ENSRNOT00000040482
ENSRNOT00000077788 ENSRNOT00000085426 |
Dpp4
|
dipeptidylpeptidase 4 |
| chr3_-_5802129 | 21.27 |
ENSRNOT00000009555
|
Sardh
|
sarcosine dehydrogenase |
| chr1_+_84304228 | 21.10 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr17_-_43504604 | 20.73 |
ENSRNOT00000083829
ENSRNOT00000066313 |
Slc17a1
|
solute carrier family 17 member 1 |
| chr16_+_54291251 | 20.62 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_+_189241593 | 19.69 |
ENSRNOT00000046025
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr4_+_163112301 | 19.61 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr7_+_94795214 | 19.28 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr17_+_69634890 | 19.21 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr4_-_23135354 | 19.12 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr7_+_29435444 | 18.91 |
ENSRNOT00000008613
|
Slc5a8
|
solute carrier family 5 member 8 |
| chr4_+_14001761 | 18.80 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr5_+_136117858 | 18.26 |
ENSRNOT00000087764
|
Tmem53
|
transmembrane protein 53 |
| chr1_+_83933942 | 17.58 |
ENSRNOT00000068690
|
Cyp2f4
|
cytochrome P450, family 2, subfamily f, polypeptide 4 |
| chr5_+_6373583 | 17.27 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr7_+_1206648 | 16.19 |
ENSRNOT00000073689
|
Pros1
|
protein S (alpha) |
| chr13_-_102790639 | 16.13 |
ENSRNOT00000003177
|
RGD1310587
|
similar to hypothetical protein FLJ14146 |
| chr16_+_72401887 | 16.09 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr5_-_124403195 | 16.02 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr2_+_26240385 | 15.76 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr14_+_6058544 | 15.71 |
ENSRNOT00000061267
|
Abcg3l3
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 3 |
| chr4_-_51199570 | 15.53 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr12_-_2568382 | 15.22 |
ENSRNOT00000035142
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr20_+_13817795 | 15.13 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr3_-_93412058 | 14.64 |
ENSRNOT00000011230
|
Cat
|
catalase |
| chr16_+_72388880 | 14.59 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr9_+_95221474 | 14.55 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_+_96639924 | 14.41 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr16_+_54332660 | 14.23 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr2_-_191294374 | 14.06 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr1_+_213676954 | 13.95 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_-_122904913 | 13.90 |
ENSRNOT00000015188
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chrM_+_9870 | 13.85 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr16_+_50049828 | 13.66 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr4_-_176294997 | 13.58 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr5_-_78376032 | 13.43 |
ENSRNOT00000075916
|
Alad
|
aminolevulinate dehydratase |
| chr13_-_104080631 | 13.25 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr18_-_48384645 | 13.21 |
ENSRNOT00000023485
|
Ppic
|
peptidylprolyl isomerase C |
| chr7_-_29171783 | 13.12 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr9_-_81400987 | 13.10 |
ENSRNOT00000035277
|
Tns1
|
tensin 1 |
| chrM_+_7758 | 12.83 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr10_+_56662561 | 12.80 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr19_+_60017746 | 12.72 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr1_+_213686046 | 12.52 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chrM_+_11736 | 12.49 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_+_248649441 | 12.43 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr10_+_14105750 | 11.99 |
ENSRNOT00000090552
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr5_-_159946446 | 11.97 |
ENSRNOT00000089184
|
Clcnka
|
chloride voltage-gated channel Ka |
| chr6_-_42630983 | 11.91 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr15_+_18322327 | 11.89 |
ENSRNOT00000009650
|
Fam3d
|
family with sequence similarity 3, member D |
| chr1_+_277068761 | 11.86 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr14_+_77067503 | 11.64 |
ENSRNOT00000085275
|
Slc2a9
|
solute carrier family 2 member 9 |
| chrM_+_10160 | 11.57 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_-_193165736 | 11.45 |
ENSRNOT00000065884
|
Lce1l
|
late cornified envelope 1L |
| chrM_+_7919 | 11.32 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr1_-_237910012 | 11.00 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr1_-_226152524 | 10.98 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr16_-_18766174 | 10.97 |
ENSRNOT00000084813
|
Sftpd
|
surfactant protein D |
| chr4_-_13878126 | 10.87 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chr9_-_91683468 | 10.75 |
ENSRNOT00000041550
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr18_-_58423196 | 10.75 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chrX_+_68771100 | 10.74 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr6_+_47940183 | 10.56 |
ENSRNOT00000011951
|
Adi1
|
acireductone dioxygenase 1 |
| chrM_+_5323 | 10.52 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr11_+_86715981 | 10.09 |
ENSRNOT00000050269
|
Comt
|
catechol-O-methyltransferase |
| chr17_+_2165906 | 10.02 |
ENSRNOT00000047043
|
RGD1563581
|
similar to S100 calcium binding protein A11 (calizzarin) |
| chr2_+_187162017 | 9.82 |
ENSRNOT00000080693
|
Insrr
|
insulin receptor-related receptor |
| chr3_-_142752325 | 9.77 |
ENSRNOT00000006200
|
Thbd
|
thrombomodulin |
| chr4_-_58875753 | 9.73 |
ENSRNOT00000016991
|
Podxl
|
podocalyxin-like |
| chr5_-_148392689 | 9.45 |
ENSRNOT00000018464
ENSRNOT00000080166 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr1_-_87147308 | 9.35 |
ENSRNOT00000027773
ENSRNOT00000089305 ENSRNOT00000090402 |
Actn4
|
actinin alpha 4 |
| chr7_-_118792625 | 9.26 |
ENSRNOT00000007398
|
Myh9
|
myosin, heavy chain 9 |
| chr5_-_107858104 | 9.26 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr4_+_153874852 | 9.13 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr9_+_17784468 | 9.05 |
ENSRNOT00000026831
|
Slc29a1
|
solute carrier family 29 member 1 |
| chr11_+_74057361 | 8.99 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr1_+_189870622 | 8.99 |
ENSRNOT00000075003
|
Tmem159
|
transmembrane protein 159 |
| chr1_-_32141036 | 8.87 |
ENSRNOT00000078039
|
Slc12a7
|
solute carrier family 12 member 7 |
| chr10_-_82852660 | 8.80 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chrX_+_15225645 | 8.77 |
ENSRNOT00000008648
|
Glod5
|
glyoxalase domain containing 5 |
| chr16_+_72268943 | 8.74 |
ENSRNOT00000032116
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr13_-_84795070 | 8.65 |
ENSRNOT00000076083
ENSRNOT00000029706 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr4_+_56337695 | 8.59 |
ENSRNOT00000071926
|
Lep
|
leptin |
| chrX_+_83678339 | 8.58 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr4_+_163174487 | 8.51 |
ENSRNOT00000088108
|
Clec9a
|
C-type lectin domain family 9, member A |
| chr7_+_141258584 | 8.49 |
ENSRNOT00000083570
|
Aqp6
|
aquaporin 6 |
| chr5_-_79008363 | 8.34 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chr11_+_82680253 | 8.18 |
ENSRNOT00000077119
ENSRNOT00000075512 |
Liph
|
lipase H |
| chr3_+_43255567 | 8.01 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr19_-_10596851 | 7.93 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr9_+_46997798 | 7.84 |
ENSRNOT00000087112
ENSRNOT00000082408 |
Il1r1
|
interleukin 1 receptor type 1 |
| chr5_-_135025084 | 7.62 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr10_-_63176463 | 7.58 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr1_+_11963836 | 7.32 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr6_-_2311781 | 7.31 |
ENSRNOT00000084171
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr5_-_173611202 | 7.27 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr1_-_275882444 | 7.26 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr3_-_52849907 | 7.12 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr2_-_154542557 | 7.11 |
ENSRNOT00000013392
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr17_+_32973695 | 7.05 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr5_+_14890408 | 7.04 |
ENSRNOT00000033116
|
Sox17
|
SRY box 17 |
| chr5_+_159484370 | 6.94 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr10_+_31880918 | 6.84 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr8_-_116465801 | 6.83 |
ENSRNOT00000085915
|
Sema3f
|
semaphorin 3F |
| chr1_+_100161872 | 6.81 |
ENSRNOT00000025777
|
Klk1c3
|
kallikrein 1-related peptidase C3 |
| chr14_+_23089699 | 6.77 |
ENSRNOT00000036948
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr18_-_74542077 | 6.74 |
ENSRNOT00000078519
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr19_+_9587653 | 6.71 |
ENSRNOT00000015956
|
Got2
|
glutamic-oxaloacetic transaminase 2 |
| chr9_+_98490608 | 6.64 |
ENSRNOT00000027232
|
Klhl30
|
kelch-like family member 30 |
| chr14_-_108412823 | 6.62 |
ENSRNOT00000081405
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr3_+_92403582 | 6.58 |
ENSRNOT00000064282
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_+_65112944 | 6.49 |
ENSRNOT00000083672
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr16_-_7250924 | 6.47 |
ENSRNOT00000025394
|
Stab1
|
stabilin 1 |
| chr13_-_53870428 | 6.46 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_114323542 | 6.46 |
ENSRNOT00000012121
|
Chrac1
|
chromatin accessibility complex 1 |
| chr1_-_241155537 | 6.43 |
ENSRNOT00000034216
ENSRNOT00000073493 |
Mamdc2
|
MAM domain containing 2 |
| chr1_-_234596971 | 6.39 |
ENSRNOT00000017892
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr1_-_175796040 | 6.36 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr7_+_117326279 | 6.30 |
ENSRNOT00000050522
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr1_+_224824799 | 6.20 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr2_+_93712992 | 6.19 |
ENSRNOT00000059326
|
Fabp12
|
fatty acid binding protein 12 |
| chr10_-_98294522 | 6.17 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr13_-_89242443 | 6.17 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chr11_-_67037115 | 6.15 |
ENSRNOT00000003137
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr1_+_168546059 | 6.11 |
ENSRNOT00000086124
|
Olr101
|
olfactory receptor 101 |
| chr5_-_33182715 | 5.97 |
ENSRNOT00000044483
|
Maged2
|
MAGE family member D2 |
| chr10_-_31790263 | 5.96 |
ENSRNOT00000059468
|
Timd2
|
T-cell immunoglobulin and mucin domain containing 2 |
| chr5_-_78378633 | 5.96 |
ENSRNOT00000076837
|
Alad
|
aminolevulinate dehydratase |
| chrX_+_128416722 | 5.92 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr15_-_8914501 | 5.91 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_168519499 | 5.90 |
ENSRNOT00000045286
|
Olr98
|
olfactory receptor 98 |
| chr2_-_227207584 | 5.85 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr2_-_205451391 | 5.85 |
ENSRNOT00000036229
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr7_+_11081536 | 5.84 |
ENSRNOT00000007110
|
Ncln
|
nicalin |
| chr4_-_98305173 | 5.82 |
ENSRNOT00000010151
|
Il23r
|
interleukin 23 receptor |
| chr10_-_40525008 | 5.79 |
ENSRNOT00000016440
|
Slc36a2
|
solute carrier family 36 member 2 |
| chr10_+_75365822 | 5.75 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr6_-_107325345 | 5.72 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr15_-_23580342 | 5.66 |
ENSRNOT00000013331
ENSRNOT00000085767 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr11_-_70499200 | 5.64 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chrM_+_3904 | 5.62 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_+_211546560 | 5.55 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr5_+_135997052 | 5.44 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr9_+_66952720 | 5.43 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr7_+_70946228 | 5.43 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chr19_+_37675113 | 5.43 |
ENSRNOT00000024125
|
RGD1561415
|
RGD1561415 |
| chr1_-_82279145 | 5.41 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr7_+_54859104 | 5.22 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr13_-_76660043 | 5.21 |
ENSRNOT00000089332
|
Pappa2
|
pappalysin 2 |
| chr4_+_175814118 | 5.20 |
ENSRNOT00000013409
ENSRNOT00000013514 |
Slco1b2
|
solute carrier organic anion transporter family, member 1B2 |
| chr4_+_147333056 | 5.18 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr17_+_77176716 | 5.16 |
ENSRNOT00000083342
ENSRNOT00000024162 |
Optn
|
optineurin |
| chr5_+_124476168 | 5.12 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr5_-_22799349 | 5.05 |
ENSRNOT00000076204
|
Asph
|
aspartate-beta-hydroxylase |
| chr2_+_127489771 | 5.05 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chrX_-_23530003 | 5.05 |
ENSRNOT00000050819
|
Shroom2
|
shroom family member 2 |
| chr7_-_5451262 | 5.00 |
ENSRNOT00000040030
|
LOC108349829
|
olfactory receptor 6C3-like |
| chr20_+_5646097 | 4.99 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr17_-_9441526 | 4.95 |
ENSRNOT00000000146
|
Txndc15
|
thioredoxin domain containing 15 |
| chr3_-_75576520 | 4.95 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr10_+_84979717 | 4.94 |
ENSRNOT00000065555
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr20_-_3818045 | 4.92 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr14_+_84447885 | 4.91 |
ENSRNOT00000009150
|
Gatsl3
|
GATS protein-like 3 |
| chr15_+_35548971 | 4.81 |
ENSRNOT00000075732
|
Olr1294
|
olfactory receptor 1294 |
| chr20_+_7718282 | 4.80 |
ENSRNOT00000000593
|
Scube3
|
signal peptide, CUB domain and EGF like domain containing 3 |
| chr11_-_71750014 | 4.79 |
ENSRNOT00000072227
|
LOC100911042
|
uncharacterized LOC100911042 |
| chr3_-_123206828 | 4.79 |
ENSRNOT00000030192
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr14_-_71973419 | 4.75 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr7_-_139929895 | 4.74 |
ENSRNOT00000047272
|
Olr1104
|
olfactory receptor 1104 |
| chr8_-_33345379 | 4.74 |
ENSRNOT00000030115
|
AABR07073381.1
|
|
| chr7_+_13151592 | 4.70 |
ENSRNOT00000010203
|
Vom2r53
|
vomeronasal 2 receptor, 53 |
| chr1_+_99648658 | 4.69 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr12_+_30202055 | 4.69 |
ENSRNOT00000001215
|
Gusb
|
glucuronidase, beta |
| chr17_+_57074525 | 4.66 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr2_+_200572502 | 4.65 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr6_+_1147012 | 4.59 |
ENSRNOT00000006360
|
Vit
|
vitrin |
| chr5_-_115387377 | 4.59 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr9_+_97355924 | 4.55 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr5_-_173312023 | 4.54 |
ENSRNOT00000026671
|
Tas1r3
|
taste 1 receptor member 3 |
| chr7_+_54859326 | 4.53 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr10_-_15306515 | 4.53 |
ENSRNOT00000060150
|
Nhlrc4
|
NHL repeat containing 4 |
| chrX_-_13116743 | 4.49 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr10_-_56531483 | 4.47 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr3_-_75923797 | 4.46 |
ENSRNOT00000073336
|
Olr587
|
olfactory receptor 587 |
| chrX_-_18096938 | 4.44 |
ENSRNOT00000042670
|
LOC100912524
|
spindlin-2B-like |
| chr2_-_140464607 | 4.42 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr9_-_81469274 | 4.39 |
ENSRNOT00000070827
|
Cxcr1
|
C-X-C motif chemokine receptor 1 |
| chr4_+_145195070 | 4.29 |
ENSRNOT00000010723
|
Mtmr14
|
myotubularin related protein 14 |
| chrX_+_65226748 | 4.26 |
ENSRNOT00000076181
|
Msn
|
moesin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 9.9 | 39.7 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 8.1 | 24.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 7.9 | 23.6 | GO:0006116 | NADH oxidation(GO:0006116) |
| 7.6 | 22.7 | GO:0061744 | motor behavior(GO:0061744) |
| 7.4 | 29.6 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 7.0 | 21.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 5.9 | 17.6 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 5.5 | 27.3 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 4.3 | 8.6 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) |
| 4.2 | 33.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 4.0 | 12.0 | GO:0030091 | protein repair(GO:0030091) |
| 3.8 | 19.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 3.4 | 20.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 3.4 | 13.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 3.3 | 13.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 3.3 | 9.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 3.2 | 19.4 | GO:0010266 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 3.1 | 40.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 3.1 | 9.3 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 3.0 | 9.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 3.0 | 12.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 3.0 | 11.9 | GO:1904975 | response to bleomycin(GO:1904975) |
| 2.8 | 11.0 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 2.7 | 10.8 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 2.7 | 16.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 2.7 | 21.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 2.6 | 7.8 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 2.5 | 10.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 2.4 | 7.3 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 2.4 | 7.3 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 2.4 | 19.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 2.3 | 7.0 | GO:0061010 | inner cell mass cellular morphogenesis(GO:0001828) endodermal cell fate determination(GO:0007493) gall bladder development(GO:0061010) |
| 2.3 | 9.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 2.2 | 8.8 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 2.2 | 11.0 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.1 | 12.7 | GO:0003383 | apical constriction(GO:0003383) |
| 2.0 | 13.9 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.9 | 5.7 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.9 | 7.6 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.8 | 5.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.8 | 3.6 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.8 | 10.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.7 | 8.7 | GO:0070189 | tryptophan catabolic process to kynurenine(GO:0019441) kynurenine metabolic process(GO:0070189) |
| 1.7 | 5.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 1.7 | 32.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.6 | 14.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.6 | 19.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 1.6 | 8.0 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 1.6 | 4.8 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 1.6 | 15.6 | GO:0009650 | UV protection(GO:0009650) |
| 1.5 | 4.4 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 1.4 | 5.8 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 1.4 | 7.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.4 | 4.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.4 | 9.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.3 | 15.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 1.3 | 13.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.3 | 40.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 1.3 | 6.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.3 | 21.8 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.2 | 6.2 | GO:0031427 | urate transport(GO:0015747) response to methotrexate(GO:0031427) |
| 1.1 | 6.8 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.1 | 3.4 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 1.1 | 9.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 1.1 | 6.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.1 | 25.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.1 | 3.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 1.0 | 5.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 1.0 | 5.9 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.0 | 20.9 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.9 | 2.8 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.9 | 10.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 25.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.9 | 3.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.9 | 24.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.9 | 4.3 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.8 | 2.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.8 | 2.5 | GO:1901740 | skeletal myofibril assembly(GO:0014866) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.8 | 6.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 3.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.8 | 16.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.8 | 2.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.7 | 6.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.7 | 4.5 | GO:0045901 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.7 | 13.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.7 | 8.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 9.3 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.7 | 2.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.7 | 4.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.7 | 2.0 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.6 | 11.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.6 | 6.4 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.6 | 3.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.6 | 4.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.6 | 2.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.5 | 11.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.5 | 2.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.5 | 1.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.5 | 4.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 16.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.5 | 5.2 | GO:1904417 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.5 | 1.4 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.5 | 1.4 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.5 | 6.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 6.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.5 | 2.7 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.4 | 5.8 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.4 | 1.3 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.4 | 2.2 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 | 6.0 | GO:0097286 | iron ion import(GO:0097286) |
| 0.4 | 6.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.4 | 3.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.4 | 9.8 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 5.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 3.8 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.3 | 24.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 19.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.3 | 3.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 3.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 2.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 3.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 10.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 1.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.3 | 7.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 2.9 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.3 | 2.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 3.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 | 1.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 | 2.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 3.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.4 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 2.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 5.0 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 4.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 8.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 10.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.2 | 2.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 4.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.2 | 0.9 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.2 | 1.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 1.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 2.4 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.2 | 0.5 | GO:0048749 | compound eye development(GO:0048749) |
| 0.2 | 2.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 7.6 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.2 | 1.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 0.7 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 4.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 21.4 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 1.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 4.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.6 | GO:0015755 | fructose transport(GO:0015755) cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 2.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 7.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 3.5 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 31.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 4.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.4 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 2.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 3.9 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 1.1 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.1 | 5.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 2.7 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 5.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 3.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 135.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 5.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 5.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 1.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 4.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 9.4 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 2.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 7.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 4.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.9 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 3.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 3.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0090166 | positive regulation of protein glycosylation(GO:0060050) Golgi disassembly(GO:0090166) spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 48.6 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.1 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 3.5 | GO:0031016 | pancreas development(GO:0031016) |
| 0.1 | 4.8 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.1 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 3.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 0.7 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 4.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 3.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.6 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 2.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 2.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 5.2 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.1 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.7 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 50.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 6.3 | 31.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 3.4 | 27.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 3.2 | 22.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.9 | 8.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 2.7 | 24.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 2.4 | 11.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 2.3 | 9.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.3 | 6.9 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 2.1 | 12.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.7 | 45.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 1.6 | 24.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 1.6 | 14.4 | GO:0042627 | chylomicron(GO:0042627) |
| 1.5 | 6.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.2 | 8.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 11.0 | GO:0031313 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 1.2 | 13.1 | GO:0032982 | myosin filament(GO:0032982) |
| 1.1 | 55.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.1 | 13.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.0 | 13.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.0 | 3.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.0 | 9.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 4.5 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.9 | 5.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.7 | 2.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.7 | 10.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 39.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.7 | 7.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.6 | 5.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 11.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 5.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.5 | 2.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.5 | 7.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 9.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 10.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 14.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 7.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 28.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 6.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 4.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 2.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 4.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 2.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 14.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 2.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 24.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 25.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 6.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 7.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 58.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 7.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 4.5 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 49.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 10.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 87.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 43.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 2.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 6.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 9.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 5.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 81.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 3.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 3.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 5.2 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 7.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 6.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 1.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 7.9 | 31.6 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 6.5 | 19.4 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 6.0 | 24.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 4.9 | 14.6 | GO:0004046 | aminoacylase activity(GO:0004046) catalase activity(GO:0004096) |
| 4.8 | 14.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 4.8 | 19.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 3.9 | 15.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 3.8 | 18.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.4 | 40.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 3.4 | 13.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 3.3 | 39.8 | GO:0019864 | IgG binding(GO:0019864) |
| 3.2 | 19.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 3.1 | 9.3 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 3.0 | 12.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 2.9 | 8.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.7 | 11.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 2.6 | 7.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 2.5 | 7.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 2.5 | 29.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 2.5 | 19.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 2.3 | 6.9 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 2.3 | 22.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 2.2 | 40.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 2.2 | 11.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.2 | 8.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 2.2 | 6.5 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 2.1 | 20.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 2.1 | 6.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.9 | 5.8 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 1.9 | 51.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.9 | 13.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.8 | 9.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.8 | 17.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.6 | 4.9 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.6 | 21.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 1.6 | 4.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.5 | 9.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.5 | 4.4 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 1.5 | 5.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 1.4 | 9.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.4 | 21.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 1.3 | 41.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.3 | 11.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.3 | 13.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.2 | 5.0 | GO:0043533 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.2 | 7.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.2 | 14.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.2 | 9.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.2 | 10.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.2 | 15.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 1.2 | 5.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.2 | 13.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.0 | 5.2 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 1.0 | 13.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.0 | 3.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.9 | 2.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.9 | 4.6 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.9 | 4.5 | GO:0033041 | sweet taste receptor activity(GO:0033041) |
| 0.9 | 2.7 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.9 | 3.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.9 | 3.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.9 | 3.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.9 | 5.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.8 | 16.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.8 | 19.7 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.8 | 6.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.8 | 9.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.8 | 2.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.7 | 5.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.7 | 2.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.7 | 12.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 16.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.7 | 9.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 3.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 2.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.6 | 10.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.6 | 8.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.6 | 4.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.5 | 11.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.5 | 5.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 18.8 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.5 | 1.6 | GO:0050497 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.5 | 6.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 1.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.5 | 10.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.5 | 8.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.4 | 10.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 5.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 4.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.4 | 9.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 7.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 2.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 7.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.4 | 6.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 4.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 2.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 12.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 3.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 3.8 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 9.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.3 | 2.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 2.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.3 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 1.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 28.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 3.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 2.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 7.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 1.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 5.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 2.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 5.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 18.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.2 | 0.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 6.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 7.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 3.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 18.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 65.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 4.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 0.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 3.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 29.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 11.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 6.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 5.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 69.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.5 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 4.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.6 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 6.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 6.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 4.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 20.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 2.3 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 7.2 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 1.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.2 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.6 | PID FOXO PATHWAY | FoxO family signaling |
| 1.0 | 15.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 29.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 2.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 50.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 3.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 5.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 19.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.3 | 12.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 9.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 11.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 5.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 5.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 5.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 4.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 6.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 3.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 8.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 34.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 66.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.6 | 31.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 1.5 | 14.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.3 | 40.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 1.2 | 29.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.1 | 19.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.1 | 15.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.1 | 11.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.0 | 3.9 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 1.0 | 8.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.0 | 30.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.9 | 5.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.8 | 12.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.8 | 45.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.8 | 14.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 8.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 9.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 12.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.6 | 2.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 57.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.6 | 6.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 19.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.6 | 10.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 8.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 6.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 6.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 6.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.5 | 15.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.5 | 5.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.5 | 11.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 5.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 6.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 6.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 10.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 13.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 9.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 6.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 7.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 10.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 5.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 3.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 4.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 5.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 12.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 6.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 5.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.1 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |