Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
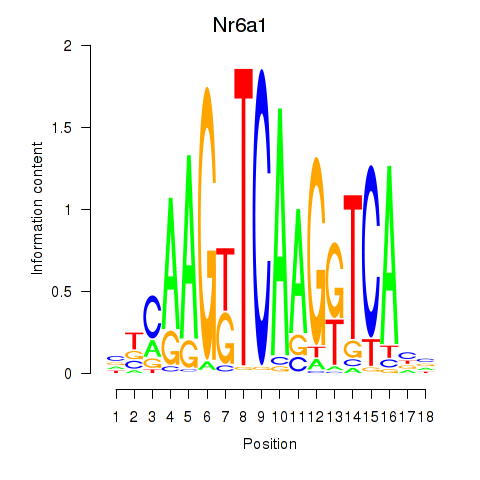
Results for Nr6a1
Z-value: 0.55
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSRNOG00000013232 | nuclear receptor subfamily 6, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr6a1 | rn6_v1_chr3_-_23066658_23066658 | 0.28 | 2.6e-07 | Click! |
Activity profile of Nr6a1 motif
Sorted Z-values of Nr6a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_40086870 | 24.61 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr10_+_71202456 | 15.00 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr1_+_72882806 | 14.56 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr1_+_199682688 | 12.45 |
ENSRNOT00000027265
ENSRNOT00000080872 |
Slc5a2
|
solute carrier family 5 member 2 |
| chr2_-_195423787 | 12.26 |
ENSRNOT00000071603
|
LOC103689947
|
selenium-binding protein 1 |
| chr20_+_21316826 | 11.85 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr2_-_196113149 | 11.65 |
ENSRNOT00000088465
|
Selenbp1
|
selenium binding protein 1 |
| chr13_+_34610684 | 11.30 |
ENSRNOT00000093019
ENSRNOT00000003280 |
Tfcp2l1
|
transcription factor CP2-like 1 |
| chr4_+_157836912 | 11.28 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr1_-_127337882 | 11.04 |
ENSRNOT00000085158
|
Aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr19_-_10653800 | 10.96 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr5_-_61077627 | 10.54 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chrX_+_15225645 | 10.38 |
ENSRNOT00000008648
|
Glod5
|
glyoxalase domain containing 5 |
| chr12_+_22153983 | 10.12 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr17_-_65955606 | 9.40 |
ENSRNOT00000067949
ENSRNOT00000023601 |
Ryr2
|
ryanodine receptor 2 |
| chr7_-_143016040 | 9.12 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr8_-_47404010 | 7.02 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr5_-_147412705 | 6.73 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr10_-_4249739 | 6.68 |
ENSRNOT00000003150
|
Snx29
|
sorting nexin 29 |
| chr3_+_139894331 | 6.41 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr8_+_131845696 | 6.20 |
ENSRNOT00000005440
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr6_+_107531528 | 5.90 |
ENSRNOT00000077555
|
Acot3
|
acyl-CoA thioesterase 3 |
| chr6_-_107325345 | 5.47 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr2_-_203494391 | 5.24 |
ENSRNOT00000021162
|
Ptgfrn
|
prostaglandin F2 receptor inhibitor |
| chr1_-_220787238 | 4.69 |
ENSRNOT00000083656
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr7_-_90318221 | 4.47 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr1_-_170404056 | 4.43 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr8_-_63150753 | 4.40 |
ENSRNOT00000078666
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr8_-_63155825 | 4.27 |
ENSRNOT00000011820
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr7_+_120755516 | 4.20 |
ENSRNOT00000056169
|
Kdelr3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr8_-_130475320 | 4.15 |
ENSRNOT00000075439
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr15_-_106844426 | 4.02 |
ENSRNOT00000015890
|
Slc15a1
|
solute carrier family 15 member 1 |
| chr4_+_56591715 | 3.90 |
ENSRNOT00000041207
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr8_+_55037750 | 3.83 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr5_-_100647727 | 3.78 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr13_-_104080631 | 3.39 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr1_+_167197549 | 3.24 |
ENSRNOT00000027427
|
Art1
|
ADP-ribosyltransferase 1 |
| chr6_-_88232252 | 3.23 |
ENSRNOT00000047597
|
Rpl10l
|
ribosomal protein L10-like |
| chr8_-_55037604 | 3.21 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr3_+_170475831 | 3.18 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr1_-_98490315 | 3.13 |
ENSRNOT00000056490
|
Cldnd2
|
claudin domain containing 2 |
| chr11_-_84633504 | 2.92 |
ENSRNOT00000052120
|
Klhl24
|
kelch-like family member 24 |
| chr4_-_179477070 | 2.90 |
ENSRNOT00000030680
|
Casc1
|
cancer susceptibility candidate 1 |
| chr1_+_21141970 | 2.74 |
ENSRNOT00000017617
|
Akap7
|
A-kinase anchoring protein 7 |
| chr12_-_30566032 | 2.66 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr7_+_2504695 | 2.66 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr4_-_65733001 | 2.64 |
ENSRNOT00000017909
|
Svopl
|
SVOP like |
| chr7_+_123102493 | 2.59 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chrX_+_40460047 | 2.57 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_+_86125316 | 2.55 |
ENSRNOT00000045930
|
Ccdc129
|
coiled-coil domain containing 129 |
| chr10_+_83081168 | 2.42 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr7_-_12519154 | 2.11 |
ENSRNOT00000093376
ENSRNOT00000077681 |
Gpx4
|
glutathione peroxidase 4 |
| chr20_-_48881119 | 2.01 |
ENSRNOT00000018892
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr1_-_71710374 | 1.94 |
ENSRNOT00000078556
ENSRNOT00000046152 |
Nlrp4
|
NLR family, pyrin domain containing 4 |
| chr5_-_157573183 | 1.81 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr20_+_3367208 | 1.70 |
ENSRNOT00000091149
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr15_+_61069581 | 1.65 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr13_+_78805347 | 1.59 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr20_-_4489281 | 1.56 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr6_-_135026449 | 1.55 |
ENSRNOT00000009444
|
RGD1305298
|
hypothetical LOC299330 |
| chr12_+_22727335 | 1.54 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr8_+_12160067 | 1.47 |
ENSRNOT00000036338
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
| chr1_-_260254600 | 1.39 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr5_-_169212170 | 1.34 |
ENSRNOT00000013385
|
Tas1r1
|
taste 1 receptor member 1 |
| chr4_+_9821541 | 1.31 |
ENSRNOT00000015733
|
Slc26a5
|
solute carrier family 26 member 5 |
| chr10_+_58875826 | 1.24 |
ENSRNOT00000020071
|
Fbxo39
|
F-box protein 39 |
| chr1_-_222350173 | 1.19 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr3_-_138708332 | 1.17 |
ENSRNOT00000010678
|
Rbbp9
|
RB binding protein 9, serine hydrolase |
| chr3_+_143109316 | 1.15 |
ENSRNOT00000006555
|
LOC257643
|
cystatin SC |
| chr9_+_40975836 | 1.12 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr7_+_2831004 | 1.10 |
ENSRNOT00000029778
|
Rnf41
|
ring finger protein 41 |
| chr5_+_156850206 | 1.08 |
ENSRNOT00000021833
|
Mul1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chrX_+_57866709 | 1.05 |
ENSRNOT00000040923
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chr7_-_118474609 | 0.94 |
ENSRNOT00000089076
|
AABR07058464.1
|
|
| chr5_-_5691794 | 0.94 |
ENSRNOT00000089152
|
AABR07046763.1
|
|
| chr1_+_193537137 | 0.91 |
ENSRNOT00000029967
|
AABR07005667.1
|
|
| chr1_+_218532045 | 0.89 |
ENSRNOT00000018848
|
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr9_-_30251388 | 0.88 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr5_-_59198650 | 0.83 |
ENSRNOT00000020958
|
Olr833
|
olfactory receptor 833 |
| chr20_+_48881194 | 0.68 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr20_-_45399694 | 0.65 |
ENSRNOT00000000715
|
Amd1
|
adenosylmethionine decarboxylase 1 |
| chrX_-_124052632 | 0.57 |
ENSRNOT00000064531
|
LOC691272
|
similar to reproductive homeobox on X chromosome, 11 |
| chr8_-_62215293 | 0.53 |
ENSRNOT00000080448
|
NEWGENE_1306267
|
phosphopantothenoylcysteine decarboxylase |
| chr5_+_144281614 | 0.51 |
ENSRNOT00000014383
|
Trappc3
|
trafficking protein particle complex 3 |
| chr1_-_84040433 | 0.45 |
ENSRNOT00000018696
|
Itpkc
|
inositol-trisphosphate 3-kinase C |
| chr5_+_140888973 | 0.44 |
ENSRNOT00000020524
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr10_-_56429748 | 0.37 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr13_-_91228901 | 0.23 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr17_+_77185053 | 0.22 |
ENSRNOT00000091561
|
Optn
|
optineurin |
| chr1_+_80954858 | 0.22 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr7_-_11724843 | 0.19 |
ENSRNOT00000084144
|
Sppl2b
|
signal peptide peptidase-like 2B |
| chr5_+_115649046 | 0.10 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_55085313 | 0.06 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr15_-_27997810 | 0.04 |
ENSRNOT00000034647
|
Olr1637
|
olfactory receptor 1637 |
| chr1_+_99781237 | 0.02 |
ENSRNOT00000025489
|
Klk5
|
kallikrein related-peptidase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr6a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0061228 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 3.7 | 11.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 3.1 | 9.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 2.1 | 10.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 2.0 | 5.9 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 1.8 | 11.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.8 | 5.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.1 | 3.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.0 | 4.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.0 | 2.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.9 | 3.8 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.9 | 11.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.9 | 2.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.8 | 3.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.7 | 11.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.7 | 2.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 2.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.7 | 2.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.6 | 4.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 2.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.4 | 1.3 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 0.4 | 2.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 1.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 12.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 1.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 5.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.3 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 11.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 11.3 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 1.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 4.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 1.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 8.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 24.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 1.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) |
| 0.1 | 3.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 3.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.6 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 4.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 4.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 8.3 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 2.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0046085 | purine nucleoside monophosphate catabolic process(GO:0009128) adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 24.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 3.6 | 14.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.1 | 4.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.1 | 3.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.7 | 2.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.5 | 12.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 11.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 3.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 2.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 9.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 22.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 12.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 4.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 9.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 7.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 13.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 1.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 13.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 10.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.3 | 26.0 | GO:0008430 | selenium binding(GO:0008430) |
| 2.4 | 9.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.3 | 4.0 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 1.3 | 2.6 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.3 | 11.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.2 | 12.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 1.2 | 11.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 1.1 | 3.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.8 | 4.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.7 | 2.0 | GO:0050567 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.5 | 2.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 11.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 5.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 3.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 1.1 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.3 | 10.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.3 | 11.0 | GO:0048020 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 5.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 1.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 4.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 2.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 15.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 8.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0002020 | protease binding(GO:0002020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 15.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 11.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 9.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 10.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 14.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 3.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 5.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 11.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 10.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 2.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 3.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 9.7 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 4.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |