Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
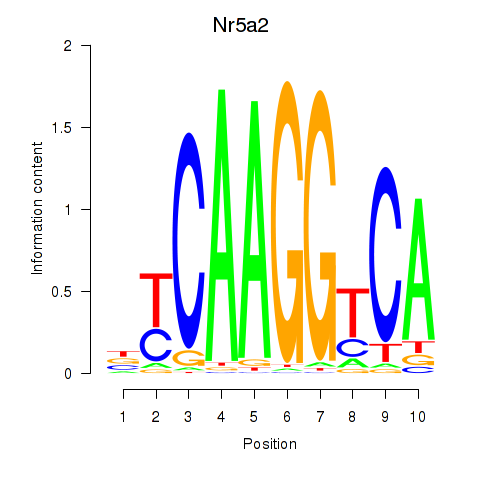
Results for Nr5a2
Z-value: 1.47
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSRNOG00000000653 | nuclear receptor subfamily 5, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr5a2 | rn6_v1_chr13_-_53870428_53870428 | -0.54 | 1.7e-25 | Click! |
Activity profile of Nr5a2 motif
Sorted Z-values of Nr5a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_33656089 | 67.79 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr8_-_130429132 | 66.37 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr9_+_82741920 | 46.61 |
ENSRNOT00000027337
|
Slc4a3
|
solute carrier family 4 member 3 |
| chr13_+_52625441 | 43.58 |
ENSRNOT00000012095
|
Tnni1
|
troponin I1, slow skeletal type |
| chr8_-_84632817 | 41.95 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr11_-_71419223 | 41.90 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr5_+_173640780 | 40.46 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr4_-_176720012 | 40.00 |
ENSRNOT00000017965
|
Ldhb
|
lactate dehydrogenase B |
| chr12_-_30566032 | 36.09 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr1_+_72882806 | 36.07 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr16_-_10941414 | 34.89 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr12_-_24365324 | 31.97 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr17_+_44758460 | 31.16 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chrX_-_40086870 | 31.06 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr13_+_52667969 | 27.92 |
ENSRNOT00000084986
ENSRNOT00000050284 |
Tnnt2
|
troponin T2, cardiac type |
| chr8_-_128266639 | 25.63 |
ENSRNOT00000064555
ENSRNOT00000066932 |
Scn5a
|
sodium voltage-gated channel alpha subunit 5 |
| chr9_+_99998275 | 25.60 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr15_+_41069507 | 25.33 |
ENSRNOT00000018533
|
C1qtnf9
|
C1q and tumor necrosis factor related protein 9 |
| chr20_+_34633157 | 24.88 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr8_+_59164572 | 24.59 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr7_+_35125424 | 24.34 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr10_+_27973681 | 24.21 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr4_+_51614676 | 21.38 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123102493 | 21.09 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chr18_+_402295 | 20.92 |
ENSRNOT00000033618
|
Fundc2
|
FUN14 domain containing 2 |
| chr4_-_16130848 | 20.82 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr3_-_60813869 | 20.75 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr10_+_45289741 | 20.61 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr4_-_16130563 | 20.16 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr9_+_43259709 | 19.70 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr18_+_83777665 | 19.68 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr10_-_62698541 | 19.33 |
ENSRNOT00000019759
|
Coro6
|
coronin 6 |
| chr17_-_44748188 | 18.40 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr5_+_118743632 | 18.39 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr3_+_132052612 | 17.80 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr2_-_119140110 | 17.71 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr2_+_119139717 | 17.39 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr17_-_44758170 | 17.20 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr7_+_12229379 | 17.05 |
ENSRNOT00000060811
|
Adamtsl5
|
ADAMTS-like 5 |
| chr4_-_157679962 | 16.83 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr8_+_116094851 | 16.56 |
ENSRNOT00000084120
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr11_+_70687500 | 16.33 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr17_-_84247038 | 16.11 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr4_-_80333326 | 15.88 |
ENSRNOT00000014058
|
LOC100363502
|
cytochrome c, somatic-like |
| chr17_-_44834305 | 15.45 |
ENSRNOT00000084303
|
Hist1h2bd
|
histone cluster 1, H2bd |
| chr7_+_117409576 | 15.33 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr13_-_103080920 | 15.31 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr13_-_47831110 | 15.08 |
ENSRNOT00000061070
|
Mapkapk2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr3_-_79728879 | 14.63 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr18_-_63825408 | 14.55 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr16_+_16949232 | 14.26 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr17_+_44520537 | 14.13 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr19_-_57699113 | 14.06 |
ENSRNOT00000026767
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr10_+_110139783 | 14.05 |
ENSRNOT00000054939
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr10_-_10881844 | 14.03 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chr3_-_3476215 | 14.01 |
ENSRNOT00000024352
|
Ubac1
|
UBA domain containing 1 |
| chr17_+_44748482 | 13.96 |
ENSRNOT00000083765
|
Hist1h2bl
|
histone cluster 1 H2B family member l |
| chr7_-_11330278 | 13.95 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr1_+_193537137 | 13.88 |
ENSRNOT00000029967
|
AABR07005667.1
|
|
| chr11_-_72109964 | 13.86 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr8_-_53901358 | 13.57 |
ENSRNOT00000047666
ENSRNOT00000042281 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr11_-_32088002 | 13.24 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr5_+_140923914 | 13.13 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr7_-_74735650 | 13.04 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr10_-_109729019 | 12.81 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chrX_+_156340925 | 12.75 |
ENSRNOT00000091810
|
Ubl4a
|
ubiquitin-like 4A |
| chr16_-_7681576 | 12.69 |
ENSRNOT00000026550
|
Colq
|
collagen like tail subunit of asymmetric acetylcholinesterase |
| chr3_+_11607225 | 12.59 |
ENSRNOT00000072685
|
St6galnac4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr6_-_136145837 | 12.34 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr19_+_26084903 | 12.21 |
ENSRNOT00000004799
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_49418965 | 12.14 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr13_-_51076852 | 12.04 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr19_+_54245950 | 12.01 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr17_+_43627930 | 11.93 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr8_+_45797315 | 11.86 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr13_-_85622314 | 11.73 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr14_-_84937725 | 11.60 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr20_+_21316826 | 11.56 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr3_+_151126591 | 11.52 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr19_-_11326139 | 11.49 |
ENSRNOT00000025669
|
Mt3
|
metallothionein 3 |
| chr8_-_119157071 | 11.44 |
ENSRNOT00000028963
|
Tmie
|
transmembrane inner ear |
| chr17_-_44520240 | 11.14 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_-_219144610 | 10.95 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr10_-_89958805 | 10.94 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr1_-_165606375 | 10.93 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chrX_+_105575759 | 10.76 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr3_+_113257688 | 10.56 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr15_+_3936786 | 10.40 |
ENSRNOT00000066163
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr19_-_10596851 | 10.17 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr19_-_37427989 | 10.09 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chrX_+_15273933 | 9.97 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr13_-_51183269 | 9.91 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr3_+_57770948 | 9.82 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr5_-_160158386 | 9.69 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chrM_+_8599 | 9.61 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr14_+_73674753 | 9.36 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr4_+_115046693 | 9.35 |
ENSRNOT00000031583
|
Bola3
|
bolA family member 3 |
| chrX_+_71155894 | 9.31 |
ENSRNOT00000076479
|
Foxo4
|
forkhead box O4 |
| chr14_-_83741969 | 9.06 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr12_+_47254484 | 8.98 |
ENSRNOT00000001556
|
Acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr1_-_170404056 | 8.91 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr6_-_135412312 | 8.91 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr14_+_58877806 | 8.89 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr8_-_55037604 | 8.70 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr18_-_63185510 | 8.69 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr6_+_1516158 | 8.58 |
ENSRNOT00000062031
|
LOC103692531
|
uncharacterized LOC103692531 |
| chr15_-_52134140 | 8.51 |
ENSRNOT00000013390
|
Polr3d
|
RNA polymerase III subunit D |
| chr4_+_7260575 | 8.46 |
ENSRNOT00000064893
|
Fastk
|
Fas-activated serine/threonine kinase |
| chr5_+_159484370 | 8.43 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chrX_-_15707436 | 8.42 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr8_+_119524039 | 8.24 |
ENSRNOT00000056050
|
Epm2aip1
|
EPM2A interacting protein 1 |
| chr8_+_55037750 | 8.23 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr1_+_226435979 | 8.17 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr10_-_15235740 | 8.07 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr6_-_50618694 | 7.88 |
ENSRNOT00000008980
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr10_-_107281235 | 7.87 |
ENSRNOT00000004196
|
Cyth1
|
cytohesin 1 |
| chr10_-_89959176 | 7.84 |
ENSRNOT00000028264
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr13_+_56262190 | 7.83 |
ENSRNOT00000032908
|
AABR07021086.1
|
|
| chr10_+_109658032 | 7.68 |
ENSRNOT00000054965
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr4_+_66405166 | 7.62 |
ENSRNOT00000077486
ENSRNOT00000067097 |
Clec2l
|
C-type lectin domain family 2, member L |
| chr20_-_48294948 | 7.62 |
ENSRNOT00000072716
|
LOC683897
|
similar to Protein C6orf203 |
| chr4_-_99746560 | 7.58 |
ENSRNOT00000012021
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr3_+_133232432 | 7.49 |
ENSRNOT00000006412
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr3_-_7141522 | 7.30 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr1_-_282251257 | 7.28 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr3_+_4083864 | 7.13 |
ENSRNOT00000006186
|
Fam69b
|
family with sequence similarity 69, member B |
| chr4_+_67378188 | 7.04 |
ENSRNOT00000030892
|
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr13_-_52413681 | 7.03 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr16_+_21050243 | 6.88 |
ENSRNOT00000064308
|
Ncan
|
neurocan |
| chr2_-_187742747 | 6.77 |
ENSRNOT00000026530
|
Bglap
|
bone gamma-carboxyglutamate protein |
| chr1_-_143535583 | 6.73 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr10_+_67862054 | 6.71 |
ENSRNOT00000031746
|
Cdk5r1
|
cyclin-dependent kinase 5 regulatory subunit 1 |
| chr16_+_1979191 | 6.67 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr3_+_79860179 | 6.60 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr14_-_18720357 | 6.60 |
ENSRNOT00000047708
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr3_+_177226417 | 6.53 |
ENSRNOT00000031328
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr1_-_78968329 | 6.47 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr3_-_8981362 | 6.35 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr1_+_31545631 | 6.33 |
ENSRNOT00000018336
|
Sdha
|
succinate dehydrogenase complex flavoprotein subunit A |
| chr8_-_130491998 | 6.31 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr7_+_121311024 | 6.23 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr1_+_222250995 | 6.15 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr1_-_175586802 | 6.09 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chr15_-_93307420 | 6.08 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr7_-_117259141 | 6.02 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr17_-_43627629 | 5.90 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr2_-_140464607 | 5.89 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr4_-_123118186 | 5.63 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr2_-_183031214 | 5.61 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr13_+_52413241 | 5.57 |
ENSRNOT00000035157
|
AABR07020999.1
|
|
| chr7_+_2504695 | 5.56 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr19_+_22569999 | 5.46 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr3_-_7796385 | 5.44 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr7_-_127058056 | 5.33 |
ENSRNOT00000022882
|
Cerk
|
ceramide kinase |
| chr7_-_119768082 | 5.25 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chr7_+_144577465 | 5.20 |
ENSRNOT00000021647
|
Hoxc10
|
homeo box C10 |
| chr10_+_84986328 | 5.16 |
ENSRNOT00000085830
ENSRNOT00000067665 |
Osbpl7
|
oxysterol binding protein-like 7 |
| chr14_-_19159923 | 5.02 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr6_+_128750795 | 4.91 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr1_+_221773254 | 4.80 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr1_+_72956026 | 4.77 |
ENSRNOT00000031462
|
Rdh13
|
retinol dehydrogenase 13 |
| chr2_+_206064179 | 4.72 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr7_+_110031696 | 4.58 |
ENSRNOT00000012753
|
Khdrbs3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr11_+_24266345 | 4.55 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr4_+_165732643 | 4.50 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr5_-_155772040 | 4.45 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr19_-_52750307 | 4.38 |
ENSRNOT00000023405
|
Zdhhc7
|
zinc finger, DHHC-type containing 7 |
| chr5_-_157573183 | 4.38 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr8_+_1502877 | 4.36 |
ENSRNOT00000034482
|
Msantd4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chr5_+_156850206 | 4.26 |
ENSRNOT00000021833
|
Mul1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr10_+_78168715 | 4.07 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr2_-_210873024 | 3.96 |
ENSRNOT00000026051
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr3_+_58164931 | 3.92 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr7_-_11777503 | 3.83 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr20_-_5618254 | 3.82 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr4_-_82209933 | 3.76 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr18_+_74299931 | 3.72 |
ENSRNOT00000078403
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog |
| chrX_-_116792707 | 3.67 |
ENSRNOT00000049482
|
Amot
|
angiomotin |
| chr7_-_12424367 | 3.64 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr10_-_67478848 | 3.55 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr3_-_123702732 | 3.52 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr7_-_121029754 | 3.50 |
ENSRNOT00000004703
|
Nptxr
|
neuronal pentraxin receptor |
| chr4_-_82295470 | 3.36 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr9_-_71830730 | 3.33 |
ENSRNOT00000019963
|
Cryga
|
crystallin, gamma A |
| chr19_-_37907714 | 3.30 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr5_+_169244778 | 3.26 |
ENSRNOT00000064841
|
Plekhg5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr4_+_145238947 | 3.23 |
ENSRNOT00000067396
ENSRNOT00000091934 |
Cpne9
|
copine family member 9 |
| chr17_-_10952441 | 3.21 |
ENSRNOT00000024580
|
Hrh2
|
histamine receptor H 2 |
| chr5_+_150001281 | 3.20 |
ENSRNOT00000034057
|
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr6_+_55085313 | 3.16 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr3_+_93909156 | 3.13 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr5_-_159962218 | 3.09 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr1_-_72674271 | 3.07 |
ENSRNOT00000023744
|
Kmt5c
|
lysine methyltransferase 5C |
| chr17_+_42133076 | 3.05 |
ENSRNOT00000031384
|
Aldh5a1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr1_+_142136452 | 2.96 |
ENSRNOT00000016445
|
Hddc3
|
HD domain containing 3 |
| chrX_+_106253355 | 2.87 |
ENSRNOT00000004252
|
LOC102552920
|
armadillo repeat-containing X-linked protein 5-like |
| chr1_+_197813963 | 2.81 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chrX_-_116792864 | 2.69 |
ENSRNOT00000090918
|
Amot
|
angiomotin |
| chr12_-_13462038 | 2.67 |
ENSRNOT00000043110
|
LOC100360439
|
ribosomal protein L36-like |
| chr1_+_224882439 | 2.65 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr5a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.6 | 67.8 | GO:1905243 | slow-twitch skeletal muscle fiber contraction(GO:0031444) response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 12.1 | 48.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 11.1 | 66.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 11.1 | 66.4 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 9.3 | 27.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 8.5 | 25.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 7.6 | 45.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 5.6 | 16.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 5.4 | 16.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 5.0 | 15.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 4.8 | 67.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 4.7 | 14.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 4.7 | 41.9 | GO:0033572 | transferrin transport(GO:0033572) |
| 4.5 | 13.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 4.4 | 43.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 4.2 | 12.7 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 4.0 | 12.0 | GO:0042323 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 3.8 | 11.5 | GO:2000295 | cadmium ion homeostasis(GO:0055073) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 3.0 | 9.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.8 | 8.4 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 2.7 | 8.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.6 | 7.9 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 2.5 | 12.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 2.5 | 10.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.5 | 19.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.3 | 18.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.2 | 6.6 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 2.1 | 4.3 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.1 | 6.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 2.0 | 12.2 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 2.0 | 27.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.9 | 15.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 1.9 | 5.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 1.8 | 5.5 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.8 | 14.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.7 | 8.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 1.7 | 6.7 | GO:0021586 | pons maturation(GO:0021586) |
| 1.6 | 13.1 | GO:0072014 | proximal tubule development(GO:0072014) |
| 1.6 | 4.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.6 | 4.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 1.6 | 6.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.5 | 7.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 1.5 | 37.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.5 | 40.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 1.5 | 40.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 1.5 | 7.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 1.4 | 5.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.4 | 12.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.3 | 3.9 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.3 | 3.8 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 1.2 | 12.1 | GO:0046684 | response to pyrethroid(GO:0046684) regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 1.2 | 9.3 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 1.0 | 8.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 1.0 | 3.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 1.0 | 18.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.9 | 4.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.9 | 5.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.9 | 8.7 | GO:0036444 | mitochondrial protein processing(GO:0034982) calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.9 | 20.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.9 | 11.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.8 | 6.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.8 | 2.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 34.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.8 | 5.6 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.8 | 65.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.7 | 9.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.7 | 6.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 76.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.7 | 23.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.7 | 31.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.7 | 4.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 2.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 9.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 8.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.6 | 12.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.6 | 6.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.5 | 4.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 1.6 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.5 | 2.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 3.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 6.6 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.5 | 6.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.5 | 2.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 5.0 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 7.3 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.4 | 6.7 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.3 | 6.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 6.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 24.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 6.3 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.3 | 14.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.3 | 8.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 3.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 2.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.7 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 7.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 18.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.2 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 8.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 13.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 31.1 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.2 | 10.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 1.8 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 3.1 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 10.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 13.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 1.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 3.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 4.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 3.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.7 | GO:0015851 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.1 | 11.4 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.1 | 3.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 3.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 5.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.1 | 4.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.3 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.1 | 1.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 3.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 7.0 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 1.9 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.5 | GO:0050798 | activated T cell proliferation(GO:0050798) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 64.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 8.3 | 24.9 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 8.2 | 24.6 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 7.8 | 23.5 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 7.0 | 41.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 6.2 | 31.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 5.6 | 67.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 5.4 | 43.6 | GO:0005861 | troponin complex(GO:0005861) |
| 3.4 | 41.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 3.2 | 12.7 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 2.9 | 8.7 | GO:0005745 | m-AAA complex(GO:0005745) |
| 2.6 | 34.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 2.3 | 11.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.2 | 6.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 2.2 | 8.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 2.1 | 16.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.9 | 37.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.8 | 29.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.6 | 80.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.6 | 154.5 | GO:0000786 | nucleosome(GO:0000786) |
| 1.6 | 7.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.6 | 17.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.4 | 37.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 1.4 | 11.5 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.4 | 39.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 1.1 | 24.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.9 | 7.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.9 | 6.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.8 | 12.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.7 | 32.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.6 | 5.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.6 | 8.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 9.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 8.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 6.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.5 | 38.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.5 | 69.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.5 | 6.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 10.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 26.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 7.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 7.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 9.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.4 | 35.7 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 4.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 6.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 7.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 22.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 9.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 6.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 23.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 8.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 5.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 11.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 6.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 7.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 17.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 6.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 17.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 37.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 6.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 7.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 6.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 64.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 13.7 | 41.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 10.0 | 40.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 9.8 | 29.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 7.5 | 15.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 6.1 | 24.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 5.1 | 25.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 5.1 | 25.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 4.9 | 19.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 4.8 | 67.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 4.7 | 41.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 4.4 | 13.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 4.2 | 16.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 4.0 | 12.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 3.7 | 18.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 3.6 | 24.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 3.0 | 9.1 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 3.0 | 11.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 2.4 | 7.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 2.4 | 12.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 2.2 | 34.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.9 | 34.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.8 | 49.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.8 | 14.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 1.7 | 24.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.7 | 8.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.6 | 6.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.4 | 25.8 | GO:0031005 | filamin binding(GO:0031005) |
| 1.3 | 15.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.2 | 3.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.2 | 3.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.1 | 30.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 1.1 | 5.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.1 | 65.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 1.0 | 11.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.0 | 3.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.0 | 4.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.0 | 7.9 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.0 | 6.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.9 | 5.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.8 | 6.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.8 | 3.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.8 | 6.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.6 | 3.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.6 | 9.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.6 | 1.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.6 | 8.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.6 | 13.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.5 | 12.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.5 | 2.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 11.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 7.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.4 | 10.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 43.8 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.4 | 7.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 9.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 6.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 5.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 6.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 3.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 7.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 13.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 4.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 6.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 89.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 136.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 29.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 2.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 6.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 10.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 5.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 12.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 8.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 6.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 5.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 9.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 3.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 8.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 7.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.6 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 74.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.7 | 24.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.7 | 25.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.6 | 15.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.6 | 10.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 12.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 32.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 6.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 18.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 20.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 3.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 3.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 12.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 6.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 7.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 4.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 4.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 4.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 22.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 7.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 10.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 11.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 77.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 2.8 | 107.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 2.6 | 54.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 2.1 | 125.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 2.0 | 52.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.4 | 41.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.4 | 24.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 18.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.2 | 25.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 1.1 | 37.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.9 | 14.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.9 | 18.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.8 | 12.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 12.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.7 | 14.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.6 | 9.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 9.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 6.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 16.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 11.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 13.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.4 | 6.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 4.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 6.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 6.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 9.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 6.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 4.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 9.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 13.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.2 | 22.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 9.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 2.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 5.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 5.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 5.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 15.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 11.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 7.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |