Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
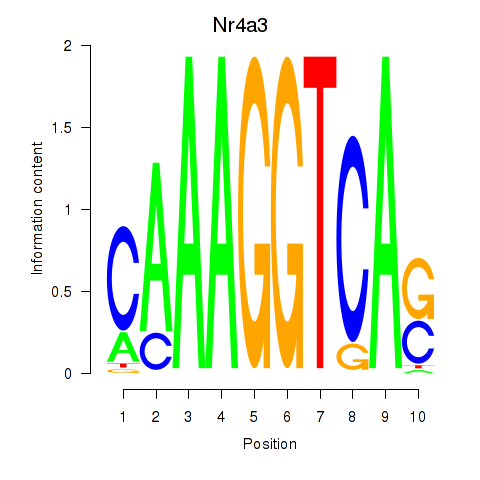
Results for Nr4a3
Z-value: 0.40
Transcription factors associated with Nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a3
|
ENSRNOG00000005964 | nuclear receptor subfamily 4, group A, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a3 | rn6_v1_chr5_+_63781801_63781801 | -0.39 | 2.5e-13 | Click! |
Activity profile of Nr4a3 motif
Sorted Z-values of Nr4a3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50531423 | 24.04 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr2_-_243407608 | 13.77 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr1_-_141579871 | 13.50 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr8_-_77398156 | 10.04 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr4_-_15505362 | 9.95 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr11_-_69201380 | 9.46 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr7_-_70661891 | 6.81 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chrX_-_139916883 | 6.61 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr10_+_71159869 | 6.46 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr8_-_119265157 | 5.92 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr3_-_92749121 | 5.05 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr11_+_83868655 | 4.85 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr15_-_14510828 | 4.77 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chrX_-_23144324 | 4.50 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr20_+_3875706 | 4.43 |
ENSRNOT00000036900
|
RT1-Ha
|
RT1 class II, locus Ha |
| chr1_-_141188031 | 3.59 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chrX_-_4945944 | 3.02 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr4_-_131694755 | 2.26 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr12_-_46718355 | 2.17 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr3_+_153398130 | 2.15 |
ENSRNOT00000068135
|
Rpn2
|
ribophorin II |
| chr1_+_196996581 | 2.15 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr16_-_85464002 | 2.14 |
ENSRNOT00000037018
|
AABR07026596.1
|
|
| chr12_+_47024442 | 2.11 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr2_-_207300854 | 1.80 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_+_167373678 | 1.78 |
ENSRNOT00000027685
ENSRNOT00000088370 |
Stim1
|
stromal interaction molecule 1 |
| chr19_-_54245855 | 1.71 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr12_-_2843772 | 1.56 |
ENSRNOT00000061750
|
Cd209c
|
CD209c molecule |
| chr4_+_176994129 | 1.39 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr16_-_37159745 | 1.33 |
ENSRNOT00000084469
|
Fbxo8
|
F-box protein 8 |
| chr15_-_106678918 | 1.13 |
ENSRNOT00000034723
|
Stk24
|
serine/threonine kinase 24 |
| chr1_+_226091774 | 1.12 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr10_-_49009156 | 1.12 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr12_-_2786628 | 1.09 |
ENSRNOT00000001348
|
RGD1564571
|
similar to DC-SIGN |
| chr10_+_88764732 | 1.02 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr20_+_3827367 | 0.61 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr4_-_145487426 | 0.34 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr12_+_6283288 | 0.10 |
ENSRNOT00000001203
|
Wdr95
|
WD40 repeat domain 95 |
| chr1_-_219544328 | 0.09 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 3.3 | 10.0 | GO:0010034 | response to acetate(GO:0010034) |
| 2.5 | 9.9 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 2.4 | 9.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.2 | 6.5 | GO:0061235 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.7 | 13.8 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.7 | 6.6 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 1.6 | 4.8 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.3 | 5.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 13.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.8 | 2.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.6 | 3.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.6 | 3.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.6 | 4.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.4 | 1.8 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 2.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 5.9 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.3 | 1.0 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) regulation of mast cell differentiation(GO:0060375) |
| 0.2 | 4.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 1.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.2 | 1.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 1.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 6.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 2.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0045988 | regulation of the force of heart contraction by chemical signal(GO:0003057) negative regulation of striated muscle contraction(GO:0045988) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 24.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.7 | 5.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.2 | 3.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 1.0 | 13.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 13.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 10.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 4.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 9.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 2.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 6.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.0 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 20.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.4 | 9.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.7 | 10.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.2 | 5.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.8 | 3.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.7 | 13.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 4.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.6 | 13.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 5.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 9.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 6.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 6.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 2.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 4.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 8.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 1.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 11.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 9.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 13.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 6.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 6.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 6.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 47.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 11.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.7 | 6.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 6.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 5.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 9.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 6.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |