Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Nr4a2
Z-value: 0.67
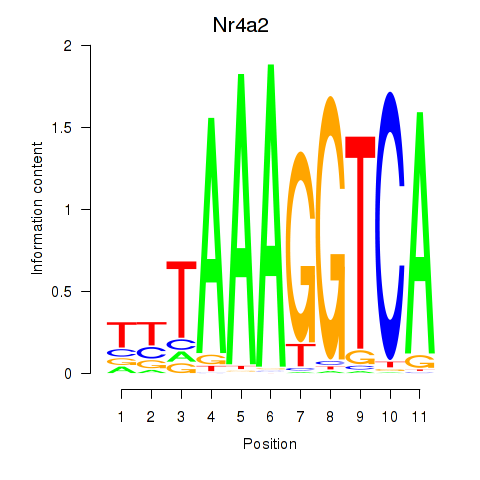
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSRNOG00000005600 | nuclear receptor subfamily 4, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | rn6_v1_chr3_-_43119159_43119159 | 0.28 | 2.9e-07 | Click! |
Activity profile of Nr4a2 motif
Sorted Z-values of Nr4a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_159775643 | 26.93 |
ENSRNOT00000010939
|
Jph2
|
junctophilin 2 |
| chr9_+_2202511 | 25.53 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr1_-_206282575 | 20.67 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr16_+_49266903 | 19.51 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chrX_+_53053609 | 18.78 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr2_+_153830586 | 17.03 |
ENSRNOT00000042576
|
Mme
|
membrane metallo-endopeptidase |
| chr10_+_57278307 | 16.12 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr9_+_64745051 | 15.32 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr16_-_22561496 | 14.56 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
| chr19_+_51317425 | 11.66 |
ENSRNOT00000019298
|
Cdh13
|
cadherin 13 |
| chr2_-_203494391 | 10.69 |
ENSRNOT00000021162
|
Ptgfrn
|
prostaglandin F2 receptor inhibitor |
| chr15_+_11298478 | 10.25 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr12_-_5822874 | 10.08 |
ENSRNOT00000075920
|
Fry
|
FRY microtubule binding protein |
| chr3_+_62648447 | 9.88 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr7_-_81592206 | 9.00 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr7_+_74047814 | 8.98 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr2_-_196304169 | 8.48 |
ENSRNOT00000028639
|
Scnm1
|
sodium channel modifier 1 |
| chr6_+_8669722 | 7.99 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr3_+_155297566 | 7.90 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr1_-_16203838 | 7.82 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr3_+_121596791 | 7.80 |
ENSRNOT00000024694
|
Ttl
|
tubulin tyrosine ligase |
| chr3_+_131351587 | 7.52 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr9_-_69953182 | 7.51 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr7_-_117288018 | 7.24 |
ENSRNOT00000091285
|
Plec
|
plectin |
| chr20_+_3827367 | 7.20 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr1_+_219000844 | 7.16 |
ENSRNOT00000022486
|
Kmt5b
|
lysine methyltransferase 5B |
| chr18_+_79813759 | 7.13 |
ENSRNOT00000021768
|
Zfp516
|
zinc finger protein 516 |
| chr20_+_3827086 | 6.62 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr11_-_69201380 | 6.51 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr2_-_250862419 | 6.43 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr7_+_53630621 | 6.33 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr17_-_80807181 | 6.19 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr9_+_88918433 | 5.90 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr19_+_49637016 | 5.84 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chrX_-_68562301 | 5.55 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr12_+_41486076 | 5.49 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr1_-_71173216 | 5.43 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr5_-_101138427 | 4.88 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr18_+_59096643 | 4.49 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chr3_+_66673071 | 4.42 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_80864846 | 4.27 |
ENSRNOT00000074412
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr9_+_94702129 | 4.25 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr2_+_196304492 | 4.23 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr14_+_83889089 | 4.06 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr7_+_35070284 | 3.74 |
ENSRNOT00000009578
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr4_-_131694755 | 3.51 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chrX_+_77263359 | 3.47 |
ENSRNOT00000077604
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr3_+_113257688 | 3.46 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr1_+_255185629 | 3.39 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chrX_+_156863754 | 3.38 |
ENSRNOT00000083611
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chrX_-_15768522 | 3.28 |
ENSRNOT00000077391
|
Foxp3
|
forkhead box P3 |
| chr18_+_61261418 | 3.09 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr3_-_72602548 | 2.84 |
ENSRNOT00000031745
|
Lrrc55
|
leucine rich repeat containing 55 |
| chr2_-_184289126 | 2.74 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr12_-_24537313 | 2.63 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr7_+_117001084 | 2.61 |
ENSRNOT00000066233
|
Zfp707l1
|
zinc finger protein 707-like 1 |
| chr7_+_144080614 | 2.61 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_-_36372436 | 2.16 |
ENSRNOT00000039827
|
Znf454
|
zinc finger protein 454 |
| chr3_-_72113680 | 2.08 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr16_+_20110148 | 1.94 |
ENSRNOT00000080146
ENSRNOT00000025312 |
Jak3
|
Janus kinase 3 |
| chr3_+_80676820 | 1.93 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr11_-_61530830 | 1.93 |
ENSRNOT00000059666
ENSRNOT00000068345 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr5_-_168004724 | 1.92 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chr4_-_157331905 | 1.74 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr10_-_49009156 | 1.68 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr8_+_114867062 | 1.58 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr2_+_51672722 | 1.56 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr3_-_2123820 | 1.47 |
ENSRNOT00000086843
|
Ehmt1
|
euchromatic histone lysine methyltransferase 1 |
| chr9_+_20213776 | 1.36 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr10_+_34975708 | 1.04 |
ENSRNOT00000064984
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr7_+_35069814 | 0.90 |
ENSRNOT00000089228
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr7_-_40316532 | 0.89 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr15_-_14510828 | 0.88 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr10_+_36590325 | 0.74 |
ENSRNOT00000085203
|
Zfp354a
|
zinc finger protein 354A |
| chr3_-_118635268 | 0.67 |
ENSRNOT00000086357
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr10_+_37069344 | 0.60 |
ENSRNOT00000073317
ENSRNOT00000090404 |
LOC103690013
|
H/ACA ribonucleoprotein complex subunit 2 |
| chr13_+_100817359 | 0.58 |
ENSRNOT00000004330
|
Tp53bp2
|
tumor protein p53 binding protein, 2 |
| chr4_-_56438465 | 0.37 |
ENSRNOT00000064007
|
Rbm28
|
RNA binding motif protein 28 |
| chr3_-_60813869 | 0.36 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr8_+_117455262 | 0.35 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr6_+_26537707 | 0.29 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chrX_+_22302703 | 0.26 |
ENSRNOT00000078617
ENSRNOT00000086952 |
Kdm5c
|
lysine demethylase 5C |
| chr1_-_81144024 | 0.19 |
ENSRNOT00000088431
|
Zfp94
|
zinc finger protein 94 |
| chr8_+_71591360 | 0.17 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr6_+_50725085 | 0.14 |
ENSRNOT00000009473
|
Slc26a3
|
solute carrier family 26 member 3 |
| chr4_+_145653602 | 0.10 |
ENSRNOT00000038964
|
Tatdn2
|
TatD DNase domain containing 2 |
| chrX_-_23144324 | 0.08 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_+_119097715 | 0.03 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 5.4 | 21.4 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 4.3 | 17.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 3.8 | 26.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 3.3 | 26.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 3.0 | 9.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 3.0 | 9.0 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 2.5 | 10.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 2.0 | 5.9 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 1.8 | 25.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.6 | 6.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.4 | 9.9 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.2 | 6.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.2 | 3.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.1 | 3.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 1.1 | 3.3 | GO:0045077 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 1.1 | 4.3 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 1.0 | 7.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.9 | 2.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.8 | 4.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.8 | 10.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 1.9 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.5 | 1.6 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 13.8 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.4 | 7.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 2.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.4 | 16.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.3 | 1.7 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.3 | 5.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 2.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 1.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.9 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 4.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 1.7 | GO:0071499 | cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 | 5.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 7.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 4.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 7.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 7.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 5.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 3.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 1.6 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.9 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 3.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 8.0 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 1.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 7.5 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.1 | 6.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 7.5 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 5.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 8.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.4 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 7.9 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.1 | GO:0033133 | NADH regeneration(GO:0006735) positive regulation of glucokinase activity(GO:0033133) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) positive regulation of hexokinase activity(GO:1903301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 26.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.7 | 16.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 14.6 | GO:0042627 | chylomicron(GO:0042627) |
| 1.0 | 18.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.8 | 5.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 2.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 6.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 25.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 7.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 7.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 1.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 9.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 6.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 7.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 11.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 17.2 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 31.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 7.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 6.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 10.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 5.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 26.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.8 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 2.7 | 16.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.4 | 7.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 2.3 | 13.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.2 | 19.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.7 | 24.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 1.6 | 6.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.5 | 5.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.2 | 3.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 1.1 | 3.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 1.1 | 4.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.0 | 3.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.9 | 18.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.9 | 6.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.6 | 1.9 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.6 | 4.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 2.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 2.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 7.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 3.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 7.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 6.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 37.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 7.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 5.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 7.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 9.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 4.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 30.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 9.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 3.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 7.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 5.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 3.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 10.1 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 6.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 8.8 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 4.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 5.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 13.7 | GO:0005102 | receptor binding(GO:0005102) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 20.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 25.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 13.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 1.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 9.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 6.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 4.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 7.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 19.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.5 | 21.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 32.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.5 | 18.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 9.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 11.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 9.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 6.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 6.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 20.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.2 | 1.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 13.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 14.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 1.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 5.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 7.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 7.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |