Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Nr3c2
Z-value: 0.75
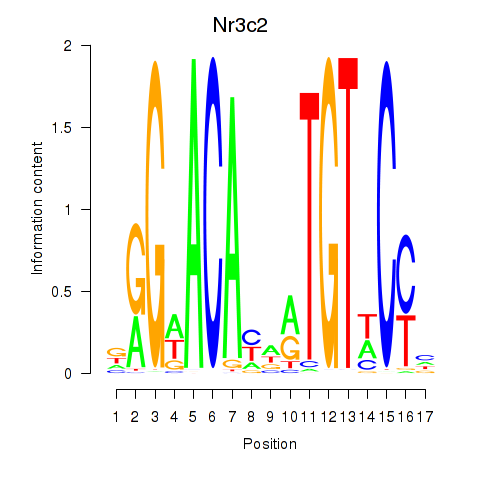
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSRNOG00000034007 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | rn6_v1_chr19_-_34752695_34752695 | 0.39 | 6.9e-13 | Click! |
Activity profile of Nr3c2 motif
Sorted Z-values of Nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_147303346 | 35.06 |
ENSRNOT00000009153
|
Hpca
|
hippocalcin |
| chr2_-_198016898 | 29.53 |
ENSRNOT00000025523
|
Car14
|
carbonic anhydrase 14 |
| chr1_+_27476375 | 29.43 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr9_+_73319710 | 24.60 |
ENSRNOT00000092485
|
Map2
|
microtubule-associated protein 2 |
| chr7_-_138483612 | 23.62 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr20_-_53777159 | 23.14 |
ENSRNOT00000073994
|
AABR07045621.1
|
|
| chr6_-_26624092 | 23.12 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr14_-_43143973 | 19.31 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr8_-_55144087 | 18.76 |
ENSRNOT00000039045
|
Dixdc1
|
DIX domain containing 1 |
| chr1_+_191344979 | 17.39 |
ENSRNOT00000023773
|
Hs3st2
|
heparan sulfate-glucosamine 3-sulfotransferase 2 |
| chrX_+_62366453 | 13.61 |
ENSRNOT00000089828
|
Arx
|
aristaless related homeobox |
| chr4_-_159079003 | 13.39 |
ENSRNOT00000026691
|
Kcna5
|
potassium voltage-gated channel subfamily A member 5 |
| chrX_+_72684329 | 10.57 |
ENSRNOT00000057644
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr15_+_52220578 | 10.50 |
ENSRNOT00000015104
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr14_-_78377825 | 9.15 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr11_-_57993548 | 8.93 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr13_+_104816371 | 8.53 |
ENSRNOT00000086497
|
AABR07022057.1
|
|
| chr3_+_95614562 | 7.21 |
ENSRNOT00000079990
|
AABR07053179.1
|
|
| chr19_+_24655626 | 6.66 |
ENSRNOT00000086690
|
AC102976.1
|
|
| chr4_-_32392007 | 6.42 |
ENSRNOT00000014946
|
Dlx5
|
distal-less homeobox 5 |
| chrX_+_153677811 | 6.41 |
ENSRNOT00000072808
|
AABR07042361.1
|
|
| chr2_+_166475070 | 6.23 |
ENSRNOT00000071111
|
AABR07011700.1
|
|
| chr1_+_91716383 | 6.19 |
ENSRNOT00000016919
|
Slc7a9
|
solute carrier family 7 member 9 |
| chr2_+_89254154 | 6.11 |
ENSRNOT00000073391
|
AABR07009221.1
|
|
| chr5_-_79899054 | 5.23 |
ENSRNOT00000074379
|
AC229945.1
|
|
| chr10_-_52483325 | 5.18 |
ENSRNOT00000083485
|
AABR07029809.1
|
|
| chr19_-_21970103 | 5.02 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr6_-_143230111 | 5.00 |
ENSRNOT00000081569
|
AABR07065844.1
|
|
| chr6_+_54438436 | 4.96 |
ENSRNOT00000091972
|
AABR07063893.1
|
|
| chr11_-_17286846 | 4.85 |
ENSRNOT00000080073
|
AABR07033318.1
|
|
| chr7_+_143707237 | 4.78 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr14_+_18231860 | 4.39 |
ENSRNOT00000003686
|
Btc
|
betacellulin |
| chr13_+_29839867 | 4.31 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chr14_-_43855745 | 4.24 |
ENSRNOT00000083155
|
AC112624.1
|
|
| chr1_-_173393390 | 4.01 |
ENSRNOT00000050657
|
Olr285
|
olfactory receptor 285 |
| chr2_+_52152536 | 3.98 |
ENSRNOT00000072691
|
AABR07008293.2
|
|
| chr3_-_76012929 | 3.91 |
ENSRNOT00000085199
|
AC111632.2
|
|
| chr3_-_2938883 | 3.89 |
ENSRNOT00000084196
|
LOC108348105
|
allergen Fel d 4-like |
| chr19_+_27603307 | 3.83 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr15_-_42271443 | 3.75 |
ENSRNOT00000085508
|
AABR07018132.1
|
|
| chr8_+_132441285 | 3.38 |
ENSRNOT00000087488
ENSRNOT00000068233 |
Lars2
|
leucyl-tRNA synthetase 2 |
| chrX_+_82710329 | 3.31 |
ENSRNOT00000074962
|
AABR07039694.1
|
|
| chr7_+_96195209 | 3.14 |
ENSRNOT00000073584
|
AABR07057961.1
|
|
| chr3_+_116072294 | 2.98 |
ENSRNOT00000071786
|
AABR07053613.1
|
|
| chr3_+_3236870 | 2.97 |
ENSRNOT00000065658
|
LOC108348105
|
allergen Fel d 4-like |
| chr7_+_143754892 | 2.90 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr7_-_123287721 | 2.81 |
ENSRNOT00000009152
|
LOC100359574
|
NHP2 non-histone chromosome protein 2-like 1-like |
| chr2_-_167607919 | 2.80 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr16_+_33373615 | 2.53 |
ENSRNOT00000079157
|
AABR07025316.1
|
|
| chr6_-_86161333 | 2.51 |
ENSRNOT00000072471
|
AABR07064590.1
|
|
| chr16_+_33373423 | 2.46 |
ENSRNOT00000091672
|
AABR07025316.1
|
|
| chr2_+_70490155 | 2.37 |
ENSRNOT00000078674
|
AABR07008681.1
|
|
| chr2_+_256609787 | 2.36 |
ENSRNOT00000047564
ENSRNOT00000088437 |
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr20_+_3364814 | 2.12 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr18_-_15284077 | 2.11 |
ENSRNOT00000046326
|
LOC498826
|
LRRGT00165 |
| chr3_-_2938663 | 1.65 |
ENSRNOT00000050294
|
LOC108348105
|
allergen Fel d 4-like |
| chr5_-_162688935 | 1.60 |
ENSRNOT00000034394
|
Aadacl4
|
arylacetamide deacetylase-like 4 |
| chr2_+_149899538 | 1.48 |
ENSRNOT00000065365
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr4_-_160605107 | 1.42 |
ENSRNOT00000090725
|
Tspan9
|
tetraspanin 9 |
| chr1_-_69016426 | 1.22 |
ENSRNOT00000093436
|
Vom2r3
|
|
| chrX_-_77529374 | 1.07 |
ENSRNOT00000078213
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr15_+_34270648 | 0.85 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr20_-_2757489 | 0.81 |
ENSRNOT00000081833
|
Btnl7
|
butyrophilin-like 7 |
| chr15_-_49467174 | 0.57 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr11_-_38274217 | 0.27 |
ENSRNOT00000002206
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr3_+_75559775 | 0.25 |
ENSRNOT00000035844
|
Pramel6
|
preferentially expressed antigen in melanoma-like 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.1 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 2.7 | 18.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 2.3 | 13.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.2 | 13.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.1 | 19.3 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 2.1 | 6.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.5 | 6.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.2 | 47.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.0 | 8.9 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.7 | 2.1 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.4 | 2.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 2.9 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.3 | 23.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 27.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 4.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 3.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 4.4 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 10.5 | GO:0017157 | regulation of exocytosis(GO:0017157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 2.9 | 35.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 2.7 | 24.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 2.8 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 19.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 1.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 23.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 8.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 6.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 16.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 10.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 17.3 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 3.3 | 13.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 2.9 | 17.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 2.3 | 29.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.5 | 6.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.1 | 24.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.0 | 13.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.9 | 2.8 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.6 | 18.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 6.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 23.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 2.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 4.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 23.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 32.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 8.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 19.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 24.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 4.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 18.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 6.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 10.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 29.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.6 | 17.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 13.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |